FIGURE 8.
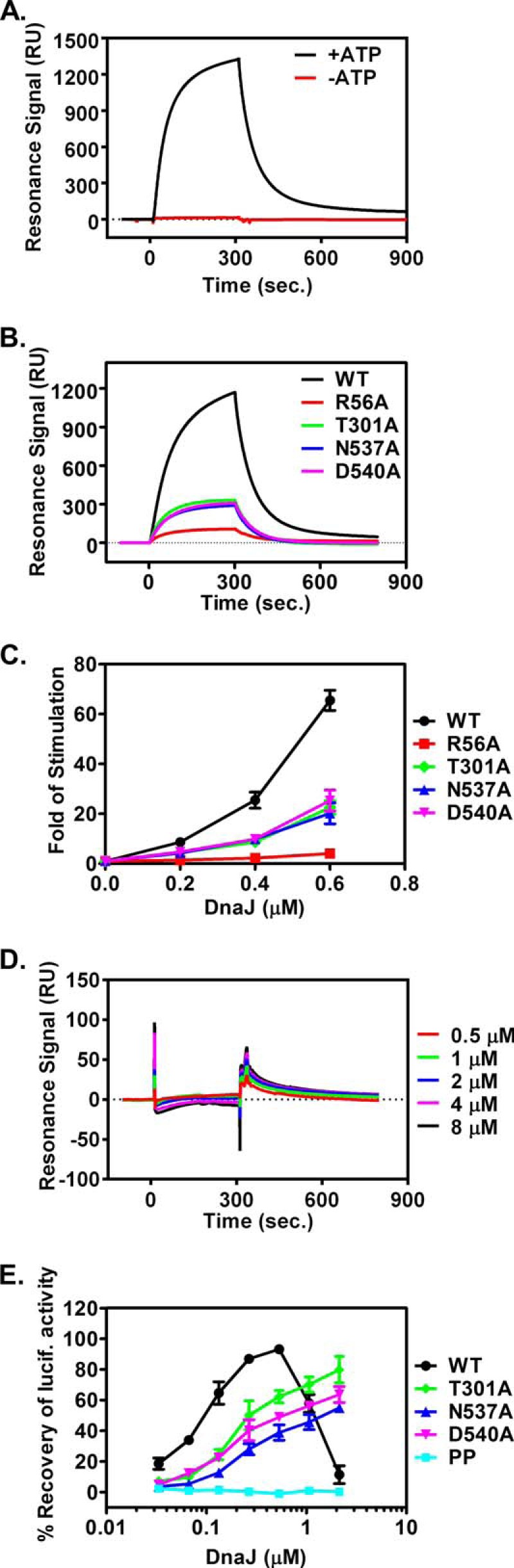
DnaJ interaction is compromised in the dimer mutant proteins. A and B, SPR analysis of the DnaK-DnaJ interaction with DnaJ immobilized. 2 μm DnaK proteins were injected over a sensor chip with DnaJ immobilized on the surface. Resonance signals after subtracting the background binding to a control channel were recorded over time. All the analyses in B were carried out in the presence of ATP. C, DnaJ stimulation of DnaK's ATPase activity. Various concentrations of DnaJ were incubated with DnaK-[32P]ATP complexes in the single-turnover ATPase assay. The hydrolysis rate, kcat, for each concentration of DnaJ was determined, and the fold of stimulation was calculated by setting the intrinsic ATPase rate as 1. D, SPR analysis with DnaK immobilized on a sensor chip. DnaJ protein was injected as analyte. The concentrations of DnaJ were labeled on the right. E, DnaJ dependence in refolding the heat-denatured firefly luciferase. Various concentrations of DnaJ were included in the refolding reactions of denatured firefly luciferase. The refolding activity was calculated by setting the undenatured luciferase activity as 100%.
