Background: RGS14 binds distinct forms of active and inactive Gα proteins through its RGS domain and GPR motif.
Results: Inactive Gαi1-GDP binding of the GPR motif does not preclude RGS action on active Gαo-GTP.
Conclusion: RGS14 simultaneously binds active Gαo and inactive Gαi1 while retaining GAP activity.
Significance: These findings clarify our understanding of how RGS14 integrates signaling by distinct G protein subunits.
Keywords: G Protein, GTPase, GTPase-activating Protein (GAP), Regulator of G Protein Signaling (RGS), Synaptic Plasticity, G Protein Regulatory (GPR) Motif, GoLoco Motif, RGS14
Abstract
RGS14 contains distinct binding sites for both active (GTP-bound) and inactive (GDP-bound) forms of Gα subunits. The N-terminal regulator of G protein signaling (RGS) domain binds active Gαi/o-GTP, whereas the C-terminal G protein regulatory (GPR) motif binds inactive Gαi1/3-GDP. The molecular basis for how RGS14 binds different activation states of Gα proteins to integrate G protein signaling is unknown. Here we explored the intramolecular communication between the GPR motif and the RGS domain upon G protein binding and examined whether RGS14 can functionally interact with two distinct forms of Gα subunits simultaneously. Using complementary cellular and biochemical approaches, we demonstrate that RGS14 forms a stable complex with inactive Gαi1-GDP at the plasma membrane and that free cytosolic RGS14 is recruited to the plasma membrane by activated Gαo-AlF4−. Bioluminescence resonance energy transfer studies showed that RGS14 adopts different conformations in live cells when bound to Gα in different activation states. Hydrogen/deuterium exchange mass spectrometry revealed that RGS14 is a very dynamic protein that undergoes allosteric conformational changes when inactive Gαi1-GDP binds the GPR motif. Pure RGS14 forms a ternary complex with Gαo-AlF4− and an AlF4−-insensitive mutant (G42R) of Gαi1-GDP, as observed by size exclusion chromatography and differential hydrogen/deuterium exchange. Finally, a preformed RGS14·Gαi1-GDP complex exhibits full capacity to stimulate the GTPase activity of Gαo-GTP, demonstrating that RGS14 can functionally engage two distinct forms of Gα subunits simultaneously. Based on these findings, we propose a working model for how RGS14 integrates multiple G protein signals in host CA2 hippocampal neurons to modulate synaptic plasticity.
Introduction
Classically defined G protein signaling begins with a heterotrimeric G protein (Gαβγ) bound to a G protein-coupled receptor (GPCR).4 GPCR activation promotes GDP release and subsequent GTP binding to activate Gα. Activation of Gα leads to dissociation/rearrangement of the heterotrimeric complex and allows Gα and Gβγ to interact with downstream effectors. Intrinsic GTP hydrolysis (GTPase) returns Gα to its basal state (Gα-GDP), where it can bind Gβγ once more and reassociate with a GPCR (1–3). G proteins are further regulated by members of the regulators of G protein signaling (RGS) family. RGS proteins contain a canonical RGS domain of ∼120 amino acids, which binds activated G proteins and acts as a GTPase-activating protein (GAP) to catalyze GTP hydrolysis and accelerate the G protein cycle (4–6).
Our previous research has revealed that RGS14 (regulator of G protein signaling 14), a member of the R12 subfamily of RGS proteins, is highly expressed in the brain (7, 8) and is a natural suppressor of CA2 hippocampal synaptic plasticity and learning and memory (9, 10). RGS14 has a unique domain structure with an N-terminal RGS domain and a C-terminal G protein regulatory (GPR; also known as GoLoco) motif. GPR motifs consist of ∼20 amino acids and bind inactive Gα-GDP subunits (11, 12), thereby targeting GPR-containing proteins to the plasma membrane (13–15). In addition to its RGS domain and GPR motif, RGS14 also contains two central tandem Raf-like Ras binding domains (RBDs), allowing RGS14 to engage H-Ras signaling pathways (16, 17).
Due to its unusual domain structure, RGS14 (together with RGS12) possesses the unique capacity for interacting with distinct forms of Gα subunits. As an RGS protein, RGS14 engages activated forms (Gα-GTP) of the Gαi/o subfamily to stimulate Gα-directed GTP hydrolysis (7, 18, 19, 55). Through its GPR motif, RGS14 selectively binds either inactive Gαi1-GDP or Gαi3-GDP to inhibit GDP dissociation and target RGS14 to the plasma membrane (13, 20, 21). However, it remains unknown which form of Gα (active or inactive) RGS14 engages first in cells and is bound to in its resting state. Potentially, RGS14 may act primarily as an RGS-GAP, recruited first by Gα-GTP following a G protein activation step and then secondarily interacting with the resulting inactive Gα-GDP to serve as a signaling complex. Alternatively, resting RGS14 may exist as a preformed complex with inactive Gα-GDP in place of Gβγ (as proposed previously (22, 23)), with its RGS domain free to interact with a second active Gα-GTP. We have previously demonstrated that recombinant RGS14 forms a stable complex with inactive Gαi at the plasma membrane (13, 24, 25). Activation of a Gαi-linked GPCR uncouples the RGS14·Gαi-GDP complex from the receptor, and the non-receptor guanosine exchange factor Ric-8A uncouples RGS14 from Gαi via the GPR motif (24, 26). Moreover, removal of the RGS domain does not prevent RGS14 localization to the plasma membrane (13), suggesting that the basal state for RGS14 is in a stable complex with Gαi-GDP.
Although we have made great strides in understanding RGS14 domain function in isolation, studies by our laboratory and others have not yet established a clear mechanism of action for how RGS14 utilizes the RGS domain and the GPR motif to integrate G protein signaling. Moreover, the structural rearrangements governing RGS14 function as an integrator of G protein signaling remain unknown. Here we explored the intramolecular dynamics of the RGS domain and the GPR motif with the goal of clarifying the RGS14 mechanism of action on Gα protein signaling. Our studies sought to distinguish between recruitment of RGS14 to the plasma membrane by the GPR motif and RGS domain. We further explored the intramolecular communication between the GPR motif and RGS domain when bound to Gα and examined whether RGS14 can engage two distinct forms of Gα subunits simultaneously. Using a variety of complementary cellular and biochemical approaches, we show that binding of active or inactive Gα subunit differentially affects RGS14 protein conformation and that RGS14 can bind both an inactive Gα-GDP and an active Gα-GDP-AlF4− subunit simultaneously to form a ternary signaling complex at the plasma membrane. Based on these findings, we propose and discuss a working model for RGS14 regulation and integration of G protein signaling at postsynaptic spines of its natural host cell, CA2 hippocampal neurons.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HeLa cells were maintained in 1× Dulbecco's modified Eagle's medium with phenol red indicator supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 mg/ml streptomycin. HEK 293 cells were maintained in 1× Dulbecco's modified Eagle's medium without phenol red indicator supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin, and 100 mg/ml streptomycin. Cells were incubated at 37 °C with 5% CO2. Transfections were carried out using polyethyleneimine (PEI) as described previously (15).
Immunofluorescence and Confocal Imaging
HeLa cells were used preferentially for confocal imaging because cell morphology allowed for better observation of the boundary between the cytosol and plasma membrane. HeLa cells were transiently transfected with 100 ng of RGS14 (pcDNA3.1-FLAG-RGS14) and/or 100 ng of Glu-Glu epitope (EE)-tagged Gα subunits (pcDNA3.1-Gαi1-EE or pcDNA3.1-Gαo-EE). Transfected cells were washed with PBS and treated with AlF4− for 10 min in Tyrode's solution (140 mm NaCl, 5 mm KCl, 1 mm MgCl2, 1 mm CaCl2, 0.37 mm NaH2PO4, 24 mm NaHCO3, 10 mm HEPES, and 0.1% glucose, pH 7.4) supplemented with 10 mm NaF, 9 mm MgCl2, and 30 μm AlCl3. Cells were then fixed at room temperature in 4% paraformaldehyde for 10 min. Excess paraformaldehyde was quenched with 200 mm Tris, pH 7.4, supplemented with 0.75% glycine. Cells were then permeabilized in 0.1% Triton X-100 for 10 min and blocked for 1 h in 8% BSA. Next, cells were incubated for 1 h at 37 °C in a 1:1000 dilution of rabbit anti-FLAG (Sigma) to detect FLAG-RGS14 and/or a 1:1000 dilution of mouse anti-EE (Covance) to detect Gα-EE in 4% BSA. Cells were then washed three times in PBS containing 0.05% Triton X-100 and placed into secondary antibodies for 30 min at 37 °C. Secondary antibodies, Alexa 488 goat anti-mouse and Alexa 594 goat anti-rabbit (Molecular Probes), were diluted 1:500 in PBS with 4% BSA. Cells were then washed twice in PBS and stained with Hoechst 33342 (1:5000) in 4% BSA for 3 min. Cells were then washed again in PBS and mounted with ProLong Gold antifade reagent (Invitrogen). Images were taken using a 60× oil immersion objective on an Olympus FV1000 confocal microscope. Images were processed, and intensity graphs were generated with ImageJ.
Bioluminescence Resonance Energy Transfer (BRET)
BRET experiments were performed in HEK 293 cells as described previously (14, 15, 26). To generate the Venus-RGS14-Luc cDNA used in the current studies, Venus was inserted at the XhoI site of the previously described phRLucN2-RGS14 (26). HEK 293 cells were transiently transfected with 5 ng of Ven-RGS14-Luc and either 0, 50, 100, 250, 500, or 750 ng of pcDNA3.1-Gαi1. For BRET experiments characterizing Gαi1(G42R) activity, HEK 293 cells were transfected with 5 ng of either RGS14-WT-Luc, Luc-RGS14-WT, or RGS14–515/516-Luc (GPR-null mutant) plasmid alone or along with 25, 50, 100, 250, or 500 ng of Gαi1(G42R)-YFP or Gαi1-WT DNA where indicated. Twenty-four hours after transfection, cells were resuspended in Tyrode's solution (140 mm NaCl, 5 mm KCl, 1 mm MgCl2, 1 mm CaCl2, 0.37 mm NaH2PO4, 24 mm NaHCO3, 10 mm HEPES, and 0.1% glucose, pH 7.4). Cells treated with AlF4− were resuspended in Tyrode's solution supplemented with 10 mm NaF, 9 mm MgCl2, and 30 μm AlCl3. After counting, 105 cells were plated into white 96-well Optiplates (PerkinElmer Life Sciences). Acceptor expression was confirmed by measuring fluorescence using the TriStar LB 941 plate reader (Berthold Technologies) with 485-nm excitation and 530-nm emission filters. BRET was monitored using 485- and 530-nm emission filters. After a 2-min application of 5 μm coelenterazine H (Nanolight Technologies), the change in BRET (ΔBRET) was calculated by dividing the 530 nm signal by the 485 nm signal (Venus/Luciferase) and subtracting the signal observed from Ven-RGS14-Luc alone. In experiments characterizing Gαi1(G42R)activity, BRET ratios (YFP/Luc) were recorded, and net BRET was calculated by subtracting the BRET signal from the luciferase alone. Data were collected using the MikroWin 2000 software and analyzed using Microsoft Excel and GraphPad Prism.
Purification of Recombinant Proteins
Full-length rat RGS14 was cloned using ligation-independent cloning into a pLIC-MBP vector (a gift from John Sondek to E. A. O.) containing a hexahistidine (H6) tag, a maltose-binding protein (MBP) tag, and a tobacco etch virus (TEV) cleavage site to generate H6-MBP-TEV-RGS14. H6-MBP-TEV-RGS14 was expressed in BL21 (DE3) Escherichia coli and purified using Ni2+ affinity chromatography. The H6-MBP tag was cleaved by treatment with purified TEV protease (1:200 TEV/RGS14) overnight at 4 °C. Pure RGS14 was isolated with size exclusion chromatography by FPLC (AKTA Purifier) utilizing tandem Superdex S75/S200 columns (GE Healthcare). Purified protein was snap frozen and stored at −80 °C. H6-Gαi1 and H6-Gαo were prepared and used as described previously (7).
Generation of G42R Mutant
An AlF4−-insensitive mutant of Gαi1 was generated by introducing a glycine to arginine mutation at amino acid 42 using the QuikChangeTM kit (Stratagene). To generate H6-Gαi1(G42R) (rat), the following oligonucleotide primers were used: forward, 5′-CTG CTG CTG CTG GGT GCT CGT GAA TCC GGG AAG AGC-3′; reverse, 5′-GCT CTT CCC GGA TTC ACG AGC ACC CAG CAG CAG CAG-3′. To generate Gαi1(G42R)-YFP (human), the following primers were used: forward, 5′-CTG CTG CTG CTC GGT GCT CGT GAA TCT GGT AAA AGT ACA ATT GTG-3′; reverse, 5′-CAC AAT TGT ACT TTT ACC AGA TTC ACG AGC ACC GAG CAG CAG CAG-3′.
Hydrogen/Deuterium Exchange (HDX) Mass Spectrometry
Solution phase amide HDX was carried out with a fully automated system as described previously (27). Briefly, 4 μl of 10 μm RGS14 was diluted to 25 μl with D2O-containing HDX buffer and incubated at 4 °C for 10, 30, 60, 900, or 3,600 s. Following on exchange, back-exchange was minimized, and the protein was denatured by dilution to 50 μl in a low pH and low temperature buffer containing 0.1% (v/v) TFA in 3 m urea (held at 1 °C). Samples were then passed across an immobilized pepsin column (prepared in house) at 50 μl min−1 (0.1% (v/v) TFA, 15 °C); the resulting peptides were trapped on a C8 trap cartridge (Hypersil Gold, Thermo Fisher). Peptides were then gradient-eluted from 4% (w/v) CH3CN to 40% (w/v) CH3CN, 0.3% (w/v) formic acid over 5 min at 2 °C across a 1 × 50-mm C18 HPLC column (Hypersil Gold, Thermo Fisher) and electrosprayed directly into an Orbitrap mass spectrometer (LTQ Orbitrap with ETD, Thermo Fisher). Peptide ion signals were confirmed if they had a MASCOT score of 20 or greater and had no ambiguous hits using a decoy (reverse) sequence in a separate experiment using a 60-min gradient. The intensity-weighted average m/z value (centroid) of each peptide's isotopic envelope was calculated with software developed in house (28) and corrected for back-exchange on an estimated 70% recovery and accounting for the known deuterium content of the on-exchange buffer. To measure the difference in exchange rates, we calculated the average percentage of deuterium uptake for RGS14 following 10, 30, 60, 900, and 3,600 s of on exchange. From this value, we subtracted the average percentage of deuterium uptake measured for the activated Gαo or inactive Gαi1-bound RGS14 complex. Negative perturbation values indicate that exchange rates are slower for these regions within RGS14 in complex with activated Gαo or inactive Gαi1. Resulting HDX data were mapped onto homologous RGS14 (PDB ID: 2JNU) or RGS10 (PDB ID: 2IHB) structures using UCSF Chimera (29). The human RGS14 RGS domain structure utilized for apo-RGS14 map was selected from the solution structure using the ensemble cluster function. Sequence alignments for RGS14 and RGS10 were performed using ClustalΩ (30, 31).
GTPase-activating Protein (GAP) Assay
Single turnover GTPase assays were performed as described previously (7, 25). Gαo was diluted in 10 mm HEPES, 5 mm EDTA, 2 mm DTT, 0.1% Lubrol and loaded with 3,500 cpm of γ-labeled [32P]GTP for 20 min at room temperature. Gαo was then cooled on ice for 5 min prior to the start of the assay. Gαo (1 μm) was added to reaction tubes containing 5 μl of 10 mm GTP and 5 μl of 1 m MgCl2 with RGS14 or RGS14·Gαi1 preformed complex. Proteins were incubated on ice for established time points and then quenched with ice-cold activated charcoal. The charcoal was pelleted, and the collected supernatant was added to scintillation vials. Released 32Pi was then measured with scintillation counting.
RESULTS
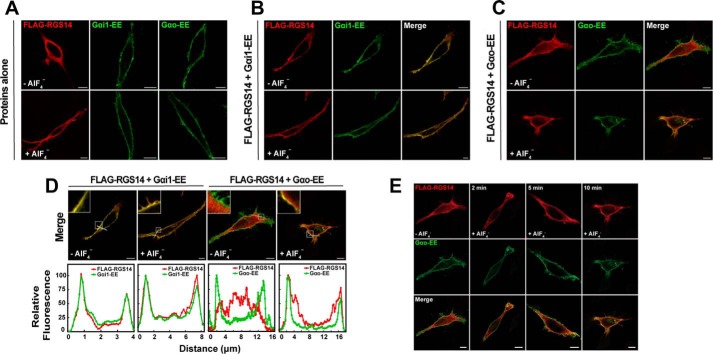
Our previous studies have indicated that native RGS14 exists both in the soluble (cytosolic) and particulate (membrane) fractions of brain lysates (7), and is localized diffusely within the soma, dendrites, and spines and at the postsynaptic density in CA2 neurons of mouse brain (8, 9). We also have shown that recombinant RGS14 can bind inactive Gα subunits at the plasma membrane through the GPR motif (13). Here we sought to explore how RGS14 subcellular localization changes in response to G protein activation. For this, we compared Gαi1, which can bind either the RGS domain or the GPR motif, and Gαo, which can only bind the RGS domain (7, 13, 18, 19, 21). Initial experiments examined subcellular localization of RGS14 in response to G protein activation with aluminum tetrafluoride (AlF4−). AlF4− activates cellular G proteins by mimicking the G protein transition state. Moreover, AlF4−-activated G proteins are the preferred binding partner for RGS domains (32–34), including RGS14. The RGS domain of RGS14 interacts with Gαo when activated with AlF4− but not Gαo activated with the non-hydrolyzable GTP analogue, GTPγS (data not shown). When expressed in HeLa cells, FLAG-RGS14 is localized diffusely within the cytosol in the absence of Gα subunits when visualized by confocal microscopy (Fig. 1A). Co-expression of EE-epitope-tagged Gαi1 (Gαi1-EE-GDP) was sufficient to recruit FLAG-RGS14 to the plasma membrane (Fig. 1, B and D). In contrast and as expected, co-expression of Gαo-EE did not recruit FLAG-RGS14 to the plasma membrane (Fig. 1, C and D). Following activation of HeLa cells with AlF4−, FLAG-RGS14 remained at the plasma membrane with Gαi1-EE but translocated from the cytosol to the plasma membrane following activation of Gαo-EE (Fig. 1D). FLAG-RGS14 translocation to the plasma membrane by Gαo-EE-AlF4− took place slowly over 10 min (Fig. 1E), probably reflecting the rate of activation of cellular Gα by AlF4−.
FIGURE 1.
RGS14 is recruited to the plasma membrane by inactive Gαi1-GDP and by active Gαo-AlF4−. A, HeLa cells were transfected with either 100 ng of FLAG-RGS14 or 100 ng of EE-tagged Gα proteins. Transfected cells were treated with AlF4− for 10 min prior to fixation for immunofluorescence and confocal microscopy as described under “Experimental Procedures.” Images are representative of three separate experiments. Scale bar, 10 μm for all panels. B, HeLa cells were co-transfected with 100 ng of FLAG-RGS14 and 100 ng of Gαi1-EE. Cells were treated with AlF4− and fixed as in A. Images are representative of three separate experiments. C, HeLa cells were co-transfected with 100 ng of FLAG-RGS14 and 100 ng of Gαo-EE. Cells were treated with AlF4− and fixed as in A. Images are representative of three separate experiments. D, intensity graphs indicating relative fluorescence from merged images in B and C. Relative fluorescence intensity of FLAG-RGS14 and either Gαi1-EE or Gαo-EE was measured at the plasma membrane as indicated by the white line in merged images. Intensity graphs were generated in ImageJ and are plotted from left to right. Insets highlight the plasma membrane in each image. E, HeLa cells were transfected with 100 ng of FLAG-RGS14 (top row) and 100 ng of EE-tagged Gαo (middle row) and treated with AlF4− for 0, 2, 5, and 10 min prior to fixation for immunofluorescence and confocal microscopy as described under “Experimental Procedures.” Merged overlay images of RGS14 and Gαo are shown (bottom row). Scale bar, 10 μm. Note that images for the zero and 10 min time points are from C.
To investigate conformational changes of RGS14 in response to G protein binding in live cells, we developed an RGS14 BRET biosensor. An acceptor Venus tag was fused to the N terminus and a donor Renilla luciferase tag to the C terminus of RGS14 (Ven-RGS14-Luc). Resonant energy transfer is dependent on the proximity and conformation of the Venus and luciferase tags; thus, conformational changes in RGS14 can alter the position of the donor and acceptor tags and register a change in the BRET signal. As seen in Fig. 2, the cytosolic Ven-RGS14-Luc biosensor exhibited basal BRET activity when expressed alone in cells. When co-expressed with inactive Gαi1, BRET activity decreased, suggesting that the BRET tags move away from one another due to binding of inactive Gαi1-GDP. In stark contrast, application of the nonspecific G protein activator AlF4− showed a marked increase in BRET signal, suggesting that the BRET tags move closer together in the presence of activated Gαi-AlF4−. These findings highlight distinct and dynamic structural rearrangements that the RGS14 polypeptide adopts in response to the binding of Gα subunits in different activation states.
FIGURE 2.
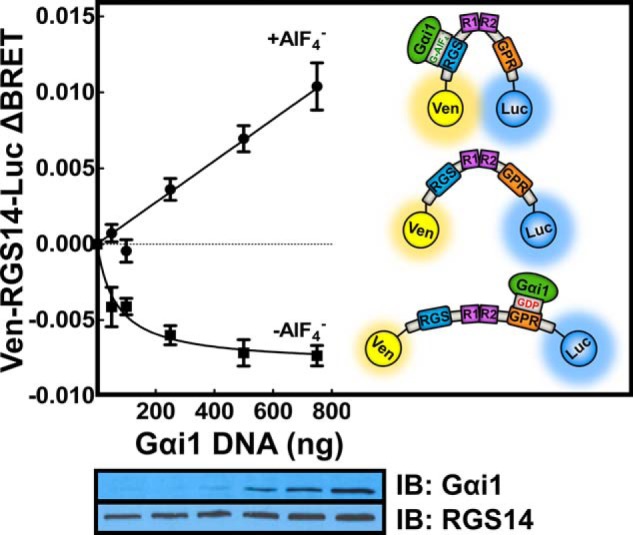
RGS14 adopts different conformations in response to binding Gα proteins in different activation states. HEK 293 cells were transfected with 5 ng of plasmid cDNA encoding Venus-RGS14-Luc alone or with 50, 100, 250, 500, or 750 ng of Gαi1. Gαi1 was activated with AlF4− for 40 min. BRET ratios were recorded, and the change in BRET (ΔBRET) was calculated by subtracting the BRET signal from Venus-RGS14-Luc alone. Average basal BRET for Venus-RGS14-Luc in the absence of AlF4− was 0.693, whereas in the presence of AlF4−, the basal BRET was 0.687. Data shown are the pooled mean ± S.E. of three separate experiments, each with triplicate determinations. The bottom panel shows representative immunoblots (IB) for Venus-RGS14-Luc and Gαi1 protein expression. The inset diagrams illustrate the interpreted dynamics of Venus-RGS14-Luc structure in response to binding Gαi1 in different activation states.
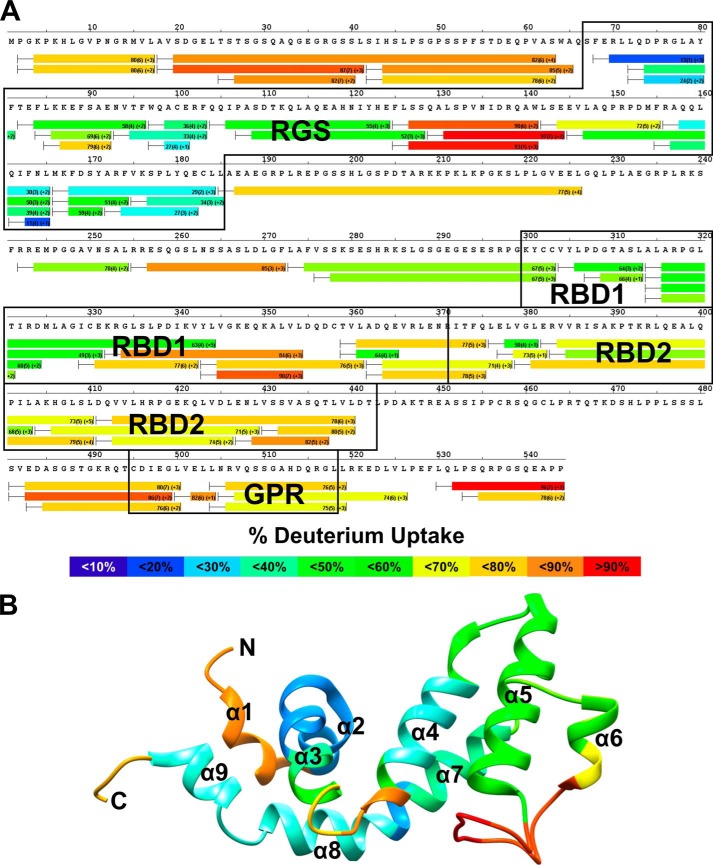
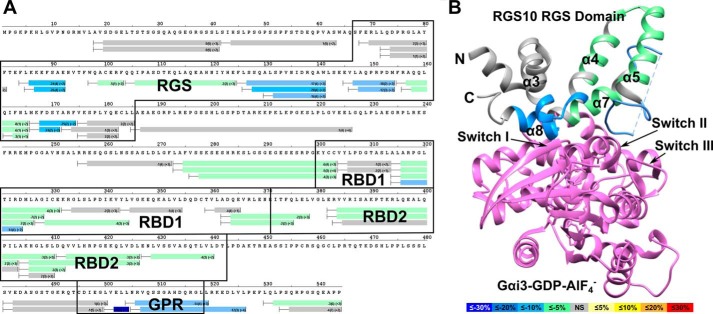
We next turned to HDX mass spectrometry to gain a better understanding of the conformational changes occurring in RGS14 in response to interactions with G proteins. HDX measures the incorporation of deuterons from heavy water (D2O) with mass spectrometry over time to probe the secondary structure. HDX has emerged as a sensitive and rapid technique to identify alterations in conformational dynamics in protein-protein or protein-ligand interactions (35–37). The HDX heat map of apo-Gαi1 (data not shown) agrees closely with previous reports of G protein structure and overall stability (38, 39), whereas the HDX heat map of apo-RGS14 indicates that RGS14 is highly dynamic in solution (Fig. 3A). High deuterium exchange was observed in both the N and C termini as well as the interdomain regions and the GPR motif. The RBDs and RGS domain were relatively more stable than the GPR motif with increased protection from solvent exchange (as determined by detection of lower levels of deuterium incorporation). The RBD1 region of RGS14 appeared to be more stable than RBD2.
FIGURE 3.
RGS14 is a highly dynamic protein. A, HDX heat map for apo-RGS14. Each bar represents an individual peptide, with the color corresponding to the average percentage of deuterium exchange over six time points (10, 30, 60, 300, 900, and 3,600 s). The numbers in the first parentheses indicate the S.D. for three replicates. The numbers in the second parentheses indicate the charge of the peptide. Residues corresponding to the RGS domain, RBDs, and GPR motif are boxed in black. The percentage deuterium exchange is indicated by the colored scale bar. B, average deuterium incorporation mapped onto the reported solution structure of human RGS14 RGS domain (PDB ID: 2JNU).
The most stable regions of apo-RGS14 as indicated by HDX are located in the RGS domain. We modeled the observed solvent exchange onto a solution structure of the RGS domain of human RGS14 (PDB ID: 2JNU) (40). As seen in Fig. 3B, residues 70–185 showed modest deuterium exchange, indicating relative stability of the domain. Within the RGS domain, peptide fragments corresponding to the α5-α6 loop (residues 127–142) showed high exchange, indicating a highly dynamic region. These results are consistent with the solution structure of the RGS domain of RGS10 (PDB ID: 2I59), a close relative of RGS14 and a member of the R12 subfamily of RGS proteins (40).
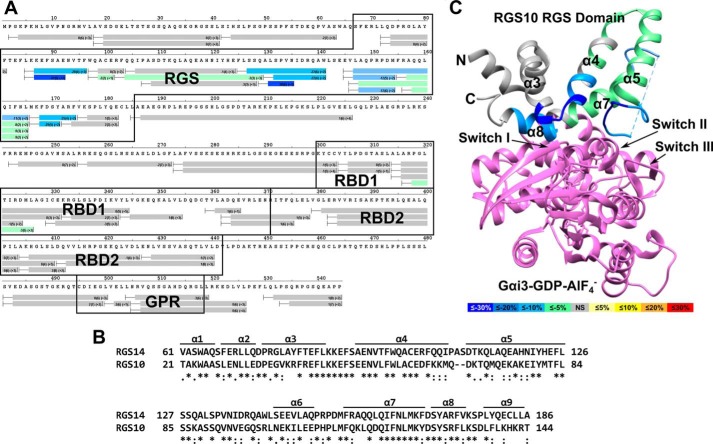
We then sought to examine the effects of G protein binding on the dynamics of RGS14 protein structure. To characterize the interaction of RGS14 with activated G proteins, we performed differential HDX with RGS14 and AlF4−-activated Gαo that binds the RGS domain but not the GPR motif (7, 18, 19). As seen in the differential HDX heat map (Fig. 4A), Gαo-AlF4− dramatically stabilizes the RGS domain, indicated by a reduced solvent exchange in RGS14 residues 87–96 (28–37%) and 127–174 (8–30%). Also, a modest yet statistically significant decrease in deuterium incorporation was observed in residues 99–126 (2–3%). The RGS domain of RGS14 demonstrates ∼50% sequence identity with the RGS domain of RGS10; thus, we modeled the changes on to a previously reported crystal structure for RGS10 in complex with AlF4−-activated Gαi3 (PDB ID: 2IHB) (40). Significant stabilization was observed in the α3-α4 and α5-α6 loops as well as α-helices 7 and 8. These regions are responsible for binding and stabilizing the switch regions of the G protein and are consistent with observed interactions between RGS domains and G proteins (41). Notably, no significant change in solvent exchange was observed in the RGS14 polypeptide outside of the RGS domain.
FIGURE 4.
Gαo activated with AlF4− binds and markedly stabilizes the RGS domain of RGS14. A, differential HDX heat map for the RGS14·Gαo-AlF4− complex. Each bar represents an individual peptide with the color corresponding to the average percentage change in deuterium exchange between apo-RGS14 and RGS14·Gαo-AlF4− over six time points (10, 30, 60, 300, 900, and 3,600 s). The numbers in the first parentheses indicate the S.D. for three replicates. The numbers in the second parentheses indicate the charge of the peptide. Residues corresponding to the RGS domain, RBDs, and GPR motif are boxed in black. Changes in deuterium exchange are indicated by the colored scale bar. B, ClustalΩ sequence alignment of rat RGS14 and human RGS10. Asterisks indicate fully conserved residues, colons indicate conservation of strongly similar properties, and periods indicate conservation of weakly similar properties. C, average percentage change in deuterium exchange levels mapped onto the crystal structure of human RGS10 bound to AlF4−-activated Gαi3 (PDB ID: 2IHB). Gαi3 is represented in purple. Differences in the percentage of deuterium exchange are indicated by the colored scale bar.
Additionally, we performed HDX experiments with RGS14 bound to inactive Gαi1-GDP. As seen in Fig. 5A, a significant decrease in deuterium incorporation (9–35%) was observed in residues 502–519, corresponding to the GPR motif. Significant changes in the stability of the RGS domain and RBD1 were also observed, indicating long range allosteric communication between RGS14 domains. As shown in Fig. 5B, allosteric stabilization was observed in residues 99–129 and 147–171 corresponding to α-helices 4 and 6/7 in the RGS domain, each showing a decrease in solvent exchange of ∼2–8%. Additionally, stabilization was observed in residues 316–328 (3–6%) and 361–378 (2–7%) in RBD1.
FIGURE 5.
Binding of Gαi1-GDP stabilizes the GPR motif and induces allosteric stabilization of other regions within RGS14. A, differential HDX heat map for the RGS14·Gαi1-GDP complex. Each bar represents an individual peptide with the color corresponding to the average percentage change in deuterium exchange between apo-RGS14 and RGS14·Gαi1 over six time points (10, 30, 60, 300, 900, and 3,600 s). The numbers in the first parentheses indicate the S.D. value for three replicates. The numbers in the second parentheses indicate the charge of the peptide. Residues corresponding to the RGS domain, RBDs, and GPR motif are boxed in black. Changes in deuterium exchange are indicated by the colored scale bar. B, average percentage change in deuterium exchange for the RGS14·Gαi1 complex mapped onto the crystal structure of human RGS10 bound to AlF4−-activated Gαi3 (PDB ID: 2IHB). Gαi3 is represented in purple. Differences in the percentage of deuterium exchange are indicated by the colored scale bar.
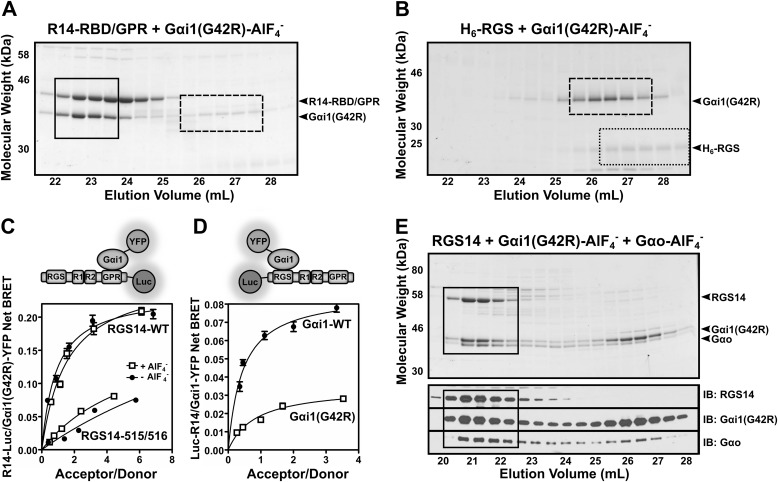
Next, we explored whether RG14 could bind two different Gα subunits simultaneously, each in different activation states, and the effects of these binding interactions on RGS14 structural stability. For these studies, we generated a Gαi1 mutant (G42R) that is insensitive to activation by AlF4− and thereby unable to bind the RGS domain but is nonetheless capable of binding the GPR motif even in the presence of AlF4− (42). Our characterization of the purified mutant protein confirmed these properties showing that Gαi1(G42R) in the presence of AlF4− readily bound the GPR motif of a truncated form of RGS14 missing the RGS domain (R14-RBD/GPR) (Fig. 6A) but failed to bind the isolated RGS domain of RGS14 (H6-RGS), again in the presence of AlF4− (Fig. 6B). We further characterized Gαi1(G42R) interactions with RGS14 in live cells using BRET analysis. For these studies, we utilized two different RGS14 BRET probes. The first contained a C-terminal tagged luciferase (RGS14-Luc) that is more sensitive to G protein binding at the GPR motif. The second contained an N-terminally tagged luciferase (Luc-RGS14) that is more sensitive to G protein binding at the RGS domain. Using the RGS14-Luc BRET probe, we found that Gαi1(G42R) interacts with the GPR motif in the presence and absence of AlF4− (Fig. 6C). A GPR-null mutant (Q515A/R516A) of RGS14-Luc showed a greatly diminished capacity for binding Gαi1(G42R), both in the presence and absence of AlF4− (Fig. 6C). BRET analysis with the N-terminally tagged Luc-RGS14 showed that Gαi1(G42R) has diminished binding to RGS14 in the presence of AlF4− compared with Gαi1-WT (Fig. 6D). Following confirmation of Gαi1(G42R) properties, we then utilized this unique experimental tool to explore the function of the RGS domain when bound to Gαi1 at the GPR motif. We preincubated purified RGS14 with purified Gαi1(G42R) and Gαo in the presence of AlF4− and then subjected these proteins to size exclusion chromatography (Fig. 6E). All three proteins co-eluted together at an elution volume consistent with a ternary protein complex, suggesting binding of Gαi1(G42R) at the GPR motif does not preclude RGS14 interactions with activated Gαo-AlF4− at the RGS domain.
FIGURE 6.
RGS14 forms a ternary complex with Gαo-AlF4− and Gαi1(G42R). A, purified truncated RGS14 missing the RGS domain (R14-RBD/GPR) was combined with H6-Gαi1(G42R) in the presence of AlF4− for 1 h at 4 °C and then run over tandem Superdex S75/S200 size exclusion columns. SDS-PAGE of collected fractions were stained with Coomassie blue. Boxed in black is the R14-RBD/GPR·Gαi1(G42R) complex, whereas free Gαi1(G42R) is indicated by the dashed box. B, purified truncated RGS14 expressing solely the RGS domain (H6-RGS) was combined with H6-Gαi1(G42R) in the presence of AlF4− for 1 h at 4 °C and then run over tandem Superdex S75/S200 size exclusion columns. SDS-PAGE of collected fractions were stained with Coomassie blue. Outlined in black dots is the free H6-RGS protein, whereas free H6-Gαi1(G42R) is indicated by the dashed box. C, HEK 293 cells were transfected with 5 ng of either RGS14-WT-Luc or RGS14-515/516-Luc (GPR-null mutant) plasmid alone or along with 25, 50, 100, 250, or 500 ng of Gαi1(G42R)-YFP DNA. Gαi1(G42R) was activated with AlF4− for 40 min. BRET ratios were recorded, and net BRET was calculated by subtracting the BRET signal from the luciferase alone. Data shown are the mean of three separate experiments, each with triplicate determinations. D, HEK 293 cells were transfected with 5 ng of Luc-RGS14 plasmid alone or along with 25, 50, 100, 250, or 500 ng of Gαi1-WT-YFP or Gαi1(G42R)-YFP DNA. Gαi1 was activated with AlF4− for 40 min. BRET ratios were recorded and net BRET was calculated by subtracting the BRET signal from the luciferase alone. Data shown are the mean of three separate experiments, each with triplicate determinations. E, purified full-length RGS14, H6-Gαo, and H6-Gαi1(G42R) proteins were incubated together in the presence of AlF4− for 1 h and then run over tandem S75/S200 size exclusion columns (top). Fractions were collected, and SDS-PAGE of collected fractions were stained with Coomassie blue. Bottom, immunoblots (IB) of each protein corresponding to fractions in the Coomassie blue-stained gel. Fractions containing the RGS14·Gαi1(G42R)·Gαo-AlF4− complex are indicated by the black box. Error bars, S.E.
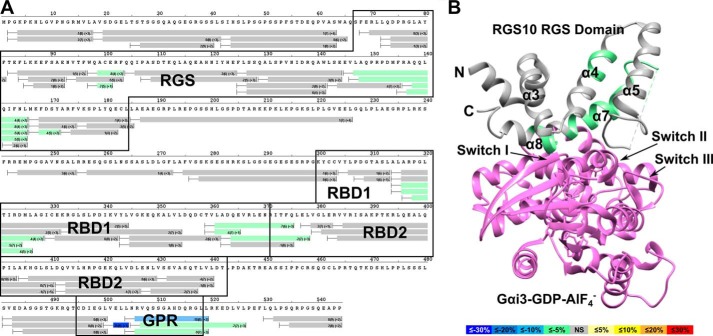
We next examined the effects of binding two different Gα subunits simultaneously at the GPR motif and the RGS domain on the RGS14 polypeptide stability using differential HDX (Fig. 7). Binding of AlF4−-activated Gαi1(G42R) and Gαo resulted in significant decreases in deuterium incorporation in both the GPR motif and RGS domain. In the RGS domain, significant decreases in solvent exchange were observed for residues 86–92 (24–26%) and 126–174 (3–25%), corresponding to the α3-α4 and α5-α6 loops as well as α-helices 7 and 8, similar to what was observed for Gαo-AlF4− binding (Fig. 4). Additional stabilizations were observed in the RGS domain in residues 99–124 (2–5%). These residues correspond to the α4-α5 loop and a large portion of the α5-helix of the RGS domain. Significant stabilization was also observed in the GPR motif. Residues 502–519 showed a 10–49% decrease of deuterium uptake, consistent with the results observed for the RGS14·Gαi1-GDP complex (Fig. 5). In addition to the RGS domain and GPR motif, stability was also observed in both Ras binding domains, as indicated by decreases in deuterium exchange of 2–14%. These results are consistent with the formation of a ternary complex.
FIGURE 7.
Simultaneous binding of Gαo-AlF4− and Gαi1(G42R) induces stability throughout RGS14. A, differential HDX heat map for the RGS14·Gαi1(G42R)·Gαo-AlF4− complex. Each bar represents an individual peptide with the color corresponding to the average percentage change in deuterium exchange between apo-RGS14 and RGS14·Gαi1(G42R)·Gαo-AlF4− over 6 time points (10, 30, 60, 300, 900, and 3,600 s). The numbers in the first parentheses indicate the S.D. for three replicates. The numbers in the second parentheses indicate the charge of the peptide. Residues corresponding to the RGS domain, RBDs, and GPR motif are outlined in black. Changes in deuterium exchange are indicated by the colored scale bar. B, average change in deuterium exchange for the RGS14·Gαi(G42R)·Gαo-AlF4− complex mapped onto the crystal structure of human RGS10 bound to AlF4−-activated Gαi3 (PDB ID: 2IHB). Gαi3 is represented in purple. Differences in percentage of deuterium exchange are indicated by the colored scale bar.
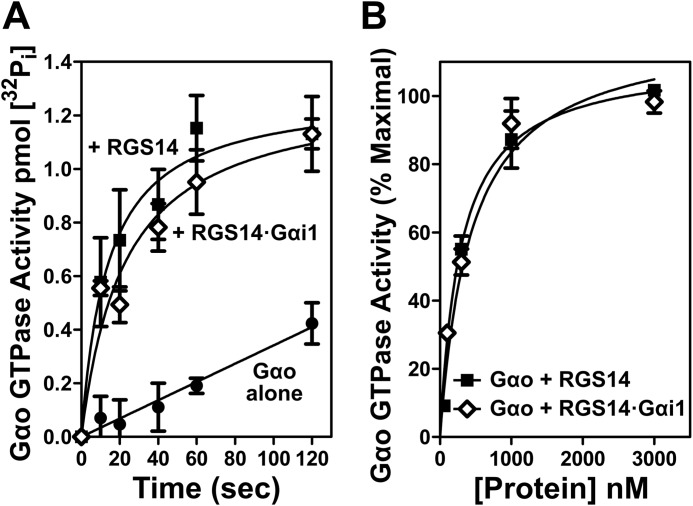
Finally, we tested whether the binding of Gαi1-GDP at the GPR motif affects the capacity of RGS14 to function as a GAP on a second Gα, Gαo-GTP. For this, we performed single turnover GTPase assays comparing RGS14 alone and a preformed complex of RGS14 bound to Gαi1-GDP (RGS14·Gαi1-GDP). As seen in Fig. 8, RGS14 alone accelerated the GTPase activity of Gαo, as expected and consistent with previous reports (7, 18, 19). The preformed RGS14·Gαi1-GDP complex also stimulated Gαo GTPase activity equally as well as RGS14 alone (Fig. 8A), and this was the case across a range of increasing protein concentrations (Fig. 8B). These results indicate that binding of Gαi1-GDP to RGS14 at the GPR motif does not alter the capacity of the RGS domain to bind Gαo-GTP and serve as a GAP.
FIGURE 8.
Gαi1-GDP binding at the GPR motif does not impede the RGS domain of RGS14 from binding Gαo-GTP and exerting GAP activity. A, purified recombinant Gαo (1 μm) was loaded with γ-labeled [32P]GTP for 20 min at room temperature, cooled on ice for 5 min, and then incubated with either 1 μm RGS14 or 1 μm RGS14·Gαi1 preformed complex for the times indicated. B, purified recombinant Gαo (1 μm) loaded with γ-labeled [32P]GTP was incubated with either 100 nm, 300 nm, 1 μm, or 3 μm RGS14 or RGS14·Gαi1 preformed complex for 20 s. In both sets of experiments, measurement of t = 0 s was performed without magnesium or RGS protein and subtracted from each condition. Each graph (A and B) is representative of three independent experiments. Error bars, S.E.
DISCUSSION
RGS14 and its close relative RGS12 are the only identified proteins that contain distinct domains that bind both active (RGS domain) and inactive (GPR motif) forms of Gα subunits. How RGS14 interacts with these two Gα in different activation states to integrate G protein signaling is unknown. Our current understanding of RGS14 biochemistry is limited to the functions of individual domains and motifs in isolation. Here we explored the intramolecular communication between the GPR motif and the RGS domains within full-length RGS14 following G protein binding. Overall, our results indicate that 1) RGS14 can exist as a preformed stable complex with Gαi1-GDP at the plasma membrane; 2) free cytosolic RGS14 can translocate to the plasma membrane in the presence of Gαo-AlF4−; 3) apo-RGS14 is a highly dynamic protein but adopts different conformations in response to binding of either active or inactive Gα; 4) Gαo-AlF4− binding stabilizes the RGS domain but does not alter the stability of other domains; 5) Gαi1-GDP binding stabilizes the GPR motif and also induces allosteric stabilization of the RGS domain and RBD1; 6) RGS14 complex formation with Gαi1-GDP at the GPR motif does not preclude binding of activated Gαo-AlF4− at the RGS domain; and 7) binding of Gαi1-GDP at the GPR motif does not disrupt the GAP activity of the RGS domain directed at a Gαo protein. Taken together, these results clarify the interdomain regulation between the RGS domain and GPR motif and demonstrate that RGS14 can functionally engage two distinct Gα subunits simultaneously. We discuss the implications of these findings for RGS14 function and propose a working model for RGS14 regulation of G protein signaling in its native environment of CA2 hippocampal neurons.
RGS14 Subcellular Localization
Our previous work has shown that native RGS14 exists as biochemically distinct subpopulations in brain, present both in the cytosol and at membranes (7, 13). Consistent with this observation, we also found that native RGS14 in hippocampal CA2 pyramidal neurons is broadly expressed in soma and dendrites and also in spine necks and postsynaptic densities (PSDs) (9). Together, these findings suggest that RGS14 exists within various subcellular compartments and is dynamically regulated. Unresolved is what form of Gα makes first contact with RGS14 within the cell (i.e. is the RGS domain first recruited to an active Gα-GTP, or is the GPR motif first recruited to an inactive Gα-GDP?). Here, we found that inactive Gαi1-GDP can preform a stable complex with RGS14 at the plasma membrane that remains there following cell-wide activation with AlF4−. In this case, we postulate that different domains of RGS14 simply swap positions with the different activation states of Gα, without falling off the membrane. By contrast, in the absence of Gαi1-GDP, RGS14 remains cytosolic yet can be recruited by activated Gαo-AlF4− to the plasma membrane via the RGS domain. Together with previous observations of RGS14 localization in brain and CA2 neurons (8, 9), our findings here suggest that RGS14 exists as distinct cellular subpopulations, either bound to the plasma membrane in complex with Gαi-GDP or recruited from the cytosol to the plasma membrane by active Gα-GTP followed by capture of the Gα-GDP resulting from GAP activity.
RGS14 Protein Conformational Changes
Following RGS14 association with Gα at the plasma membrane, an unresolved question is how Gα binding affects RGS14 protein conformation. As we previously postulated (26), RGS14 may form a clam shell-like structure, similar to other GPR scaffolding proteins (43), that closes after binding active Gα-GTP and opens upon binding inactive Gα-GDP. In support of this idea, we found here that an RGS14 biosensor (Ven-RGS14-Luc) exhibited intrinsic BRET activity that increased following binding of active Gαi1-AlF4− at the RGS domain and decreased following binding of inactive Gαi1-GDP at the GPR motif. This suggests that in the absence of Gα, apo-RGS14 exists in a semi-clam shell conformation that opens following binding of inactive Gα-GDP and closes following binding of Gα-AlF4−, thus reflecting the N and C termini of RGS14 becoming more compact. In either case, Gα binding could alter RGS14 interactions with other known RGS14 partners (e.g. active H-Ras, Rap2, or Ca2+/CaM) and/or put RGS14 in a conformation that is optimal for regulation (e.g. phosphorylation or other).
As a complement to these findings, we utilized HDX to explore the dynamic nature of purified apo-RGS14 as well as dynamic changes that might occur to RGS14 structure following binding Gα subunits. Apo-RGS14 demonstrated considerable intrinsic flexibility, particularly within the interdomain regions and at both termini (Fig. 3). As a scaffolding protein capable of integrating signals from different Gα proteins and H-Ras, RGS14 flexibility in the interdomain regions would allow for the adoption of different conformations when binding different proteins. The GPR motif exhibited considerable flexibility, consistent with a previous report showing that a short peptide corresponding to the RGS14 GPR motif utilizes an α-helix to contact the Ras-like lobe and irregular secondary structure to contact the α-helical lobe of Gαi1-GDP (40, 44). As such, many residues within this motif must remain accessible to solvent in order to bind Gα.
The tandem Ras binding domains (RBD1 and RBD2) also showed considerable flexibility. Of these, RBD1 was most stable, particularly within residues 315–330. Based on homologous RBDs (45), these residues correspond to the β2-sheet and α1-helix known to engage small GTPases, such as H-Ras and Rap2. The increased stability in RBD1 may reflect the functionality of the domain because H-Ras and Rap2 bind RBD1 but not RBD2 (17, 46). At present, there are no known binding partners of RBD2, but in order to engage other binding partners, this domain may require additional stabilization by (as yet unknown) posttranslational modification(s), an adjacent lipid bilayer, and/or ancillary binding partners.
Last, the RGS domain of apo-RGS14 showed the most stability, consistent with a folded RGS domain. Peptides corresponding to the α2-, α3-, α4-, and α7-helices were most stable and are known to contribute to the hydrophobic core in homologous RGS domain structures (41). Peptides corresponding to the α3-α4 and α5-α6 loops were least stable, reflecting the solvent-accessible G protein binding site where the RGS domain contacts and stabilizes the switch regions of the G protein to promote GTP hydrolysis (40, 41). In summary, apo-RGS14 exhibited dynamic structural properties typical of scaffolding proteins that integrate signals from many binding partners.
We also examined the effects of Gα binding on RGS14 structural stability by differential HDX. Upon binding Gαo-AlF4−, the α3-α4 and α5-α6 loops of the RGS domain show the most stabilization. Additionally, stabilization was observed in helix 7/8, regions known to interact with and stabilize G protein switch regions. These results are consistent with previous reports of G protein interactions with RGS domains (40, 41). Somewhat surprisingly, the differential HDX map for RGS14·Gαo-AlF4− did not show significant changes in other protein regions, suggesting that RGS14 interactions with activated Gα-GTP do not regulate the function of other RGS14 domains. Conversely, the differential HDX map for RGS14·Gαi1-GDP suggests that binding at the GPR motif has effects on other domain stability. Binding of Gαi1-GDP stabilized the GPR motif, as expected. However, stability was also observed in the RGS and RBD1 domains. These changes do not appear to be the result of direct protein binding to the RGS and RBD domains but rather allosteric stabilization, suggesting that the Gαi1-GDP stabilization of RGS14 could modulate Gα-GTP binding via the RGS domain and H-Ras-GTP binding at RBD1. Consistent with this observation, we previously reported that Gαi1-GDP binding at the GPR motif markedly enhanced H-Ras-GTP interactions with RGS14 (26).
To understand the implications of stabilization of the RGS domain by binding of Gαi1-GDP at the GPR motif, we modeled the stabilized regions onto a previously reported structure of RGS10 bound to Gαi3 (PDB ID: 2IHB) (40). The observed regions of stabilization were located away from the G protein binding site in α-helices 4 and 6/7. Within the residues identified, a conserved cysteine is present in the α4-helix, which has been shown to be palmitoylated in RGS4 (Cys-95) and RGS10 (Cys-66) (47). Although RGS14 has not been shown to be palmitoylated to date, the allosteric changes in the RGS domain may allow for such regulation to occur. Additionally, the changes observed in the RGS domain are proximal to identified PIP3 binding sites in other RGS proteins (48, 49), suggesting these alterations in domain stability may place RGS14 in a more favorable position to interact with the plasma membrane.
RGS14 Complex Formation with Two G Proteins
We examined whether RGS14 could simultaneously interact with an active Gαo-AlF4− at the RGS domain and an inactive Gαi1(G42R)-GDP at the GPR motif. By size exclusion chromatography (Fig. 6E), RGS14 appears to form a ternary complex with active Gαo-AlF4− and inactive Gαi1(G42R)-GDP. This finding raised two additional questions: 1) how does simultaneous binding of two proteins alter RGS14 protein conformation, and 2) does binding of Gαi1-GDP at the GPR motif affect the GAP activity of RGS14 toward an active Gαo-GTP? Our differential HDX studies indicated that binding of two Gα proteins simultaneously results in overall stability of RGS14. Somewhat surprisingly, binding of Gαi1-GDP at the GPR motif appears to not affect RGS14 GAP activity toward Gαo-GTP. A preformed RGS14·Gαi1-GDP complex retains full and unaltered capacity to directly bind Gαo-GTP and accelerate its GTPase activity by single turnover GTPase assays, irrespective of the conformational changes within the RGS domain indicated by HDX.
Proposed Working Model
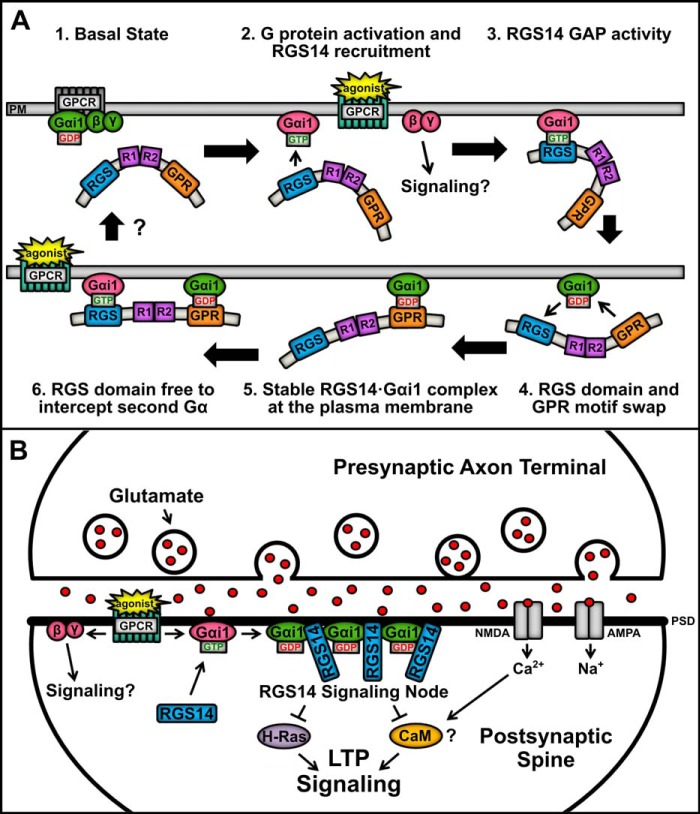
Based on our findings here and elsewhere, we propose a working model (Fig. 9) for how RGS14 integrates G protein signaling in its native environment, CA2 hippocampal neurons. Our data are most consistent with a model where RGS14 is recruited by an activated Gα-GTP and then is captured at the plasma membrane by Gα-GDP. Native RGS14 is most abundant in the cytosol (soma, dendrites, and spines) of CA2 hippocampal neurons and in extracts (soluble fraction) of whole brain (7, 9). In our proposed model, inactive RGS14 is present in the cytosol unbound to Gα. Following stimulation of a postsynaptic GPCR (Fig. 9A), RGS14 is initially recruited as a GAP to the plasma membrane by Gαi1-GTP via the RGS domain. Following GAP activity, the resulting Gα-GDP immediately binds the GPR motif, thereby capturing and anchoring RGS14 at the plasma membrane to regulate signaling pathways important for LTP. We propose that this newly formed RGS14·Gα-GDP complex can serve as a nucleating center for the recruitment and clustering of additional RGS14 at the plasma membrane within the immediate vicinity to serve as a signaling hot spot. As illustrated in Fig. 9A, the RGS domain is free to GAP one or more adjacent Gα-GTP subunits to form free Gα-GDP, each capable of recruiting RGS14 to form a cluster of RGS14·Gα-GDP complexes. Within the dendritic spines of CA2 neurons (Fig. 9B), coincident activation of AMPA/NMDA receptors and nearby Gαi-linked group II metabotropic glutamate receptors (or other GPCRs) by released glutamate may recruit a cluster of RGS14·Gα-GDP to the plasma membrane (or PSD). As we have reported previously, this complex is capable of interacting with active H-Ras-GTP to inhibit ERK signaling (17) and Ca2+/CaM (50), both key regulators of LTP and synaptic plasticity (51, 52). Recruitment of RGS14 would limit Gαi1-GTP signaling, and the newly formed RGS14·Gαi-GDP complex is now well positioned to sequester H-Ras-GTP and/or CaM activated by Ca2+ influx from NMDA receptors and thereby inhibit downstream signaling events essential for LTP.
FIGURE 9.
Proposed model showing RGS14 binding and integration of Gα-GDP and Gα-GTP signaling in cells. A, the proposed model of RGS14 signaling function proceeds clockwise from the top left. (1) In the basal resting state, Gαi1-GDP in complex with Gβγ is bound to a GPCR at the plasma membrane (PM), whereas RGS14 remains in the cytosol. (2) Agonist activation of a GPCR induces downstream signaling through Gβγ, whereas activated Gαi1-GTP recruits RGS14 to the plasma membrane. (3) RGS14 accelerates the GTPase of Gαi1, causing hydrolysis of GTP to GDP. (4) The RGS domain loses affinity for Gαi1-GDP, and the GPR motif is free to bind the newly inactivated Gαi1. (5) RGS14 forms a stable complex with inactive Gαi1-GDP at the plasma membrane, which could serve to nucleate local recruitment of other RGS14·Gαi1-GDP complexes. (6) The RGS domain is then free to intercept and “GAP” a second near Gαi1-GTP after activation of a nearby GPCR, generating a Gαi1-GDP that could recruit a second RGS14. Unresolved (?) is how the complex is regulated to return to basal state (1). B, in the dendritic spines of CA2 hippocampal neurons, RGS14 is recruited to the PSD by activated Gαi1-GTP following coincident activation of a GPCR (e.g. metabotropic glutamate receptors and others) and iGluRs (AMPA and NMDA receptors). Following RGS domain-catalyzed GTP hydrolysis, RGS14 may remain bound at the membrane by inactive Gαi1-GDP through the GPR domain, thereby concentrating RGS14 at the PSD. This serves to nucleate subsequent recruitment of a collection of RGS14·Gαi1 complexes at the PSD to form a signaling node that can intercept incoming excitatory signals promoting LTP, such as H-Ras and Ca2+/calmodulin (CaM). Clustering of inactive Gαi1-GDP in an RGS14·Gαi1 signaling node may also promote sustained signaling by activated Gβγ.
An implicit prediction of this model is that RGS14 sequestering of Gα-GDP would enhance/prolong the signaling of the Gβγ abandoned by the Gα-GDP bound to RGS14. Therefore, the purpose of this GPCR signaling event in dendrites/spines of CA2 neurons may be to nucleate a localized cluster of RGS14·Gα-GDP to serve as a signaling center, block LTP, and enhance Gβγ signaling (for as yet unknown purposes). Cellular mechanisms that regulate and reverse this complex are poorly understood at this time, although the RGS14·Gα-GDP complex is phosphorylated (53, 54) and can be regulated by a GPCR and the non-receptor guanine nucleotide exchange factor Ric8A (24, 25), suggesting possible modes of regulation for further investigation. Although speculative, this model is entirely consistent with existing data and makes predictions that are testable and under investigation. In this way, RGS14 may act to suppress LTP and synaptic plasticity in CA2 hippocampal neurons.
Acknowledgments
We thank Cindee K. Yates for generating the H6-MBP-TEV-RGS14 cDNA and for general technical assistance. We also thank Dr. Oskar Laur (Emory DNA Custom Cloning Core Facility) for generating the Venus-RGS14-Luc cDNA, Michel Bouvier (University of Montreal) for the phRLucN2 vector, and John Sondek (University of North Carolina, Chapel Hill, NC) for the pLIC-MBP vector.
This work was supported, in whole or in part, by National Institutes of Health Grants 5R01NS037112 and 5R21NS074975 (to J. R. H.) and National Institutes of Health Pharmacological Sciences Predoctoral Research Training Grant T32M008602 (to Emory University in support of N. E. B.). Additional support was provided by an Emory URC grant (to J. R. H.) and American Heart Association Predoctoral Grant 14PRE18850017 (to N. E. B.). This project was also supported in part by the Emory University Integrated Cellular Imaging Microscopy Core of the Emory Neuroscience NINDS, National Institutes of Health, Core Facilities Grant P30NS055077.
- GPCR
- G protein-coupled receptor
- RGS
- regulator of G protein signaling
- GPR
- G protein regulatory
- RBD
- Ras-binding domain
- GAP
- GTPase-activating protein
- BRET
- bioluminescence resonance energy transfer
- HDX
- hydrogen/deuterium exchange
- MBP
- maltose-binding protein
- TEV
- tobacco etch virus
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- PSD
- postsynaptic density.
REFERENCES
- 1. Gilman A. G. (1987) G proteins: transducers of receptor-generated signals. Annu. Rev. Biochem. 56, 615–649 [DOI] [PubMed] [Google Scholar]
- 2. Hepler J. R., Gilman A. G. (1992) G proteins. Trends Biochem. 17, 383–387 [DOI] [PubMed] [Google Scholar]
- 3. Hamm H. E. (1998) The many faces of G protein signaling. J. Biol. Chem. 273, 669–672 [DOI] [PubMed] [Google Scholar]
- 4. Hollinger S., Hepler J. R. (2002) Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol. Rev. 54, 527–559 [DOI] [PubMed] [Google Scholar]
- 5. Ross E. M., Wilkie T. M. (2000) GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu. Rev. Biochem. 69, 795–827 [DOI] [PubMed] [Google Scholar]
- 6. De Vries L., Zheng B., Fischer T., Elenko E., Farquhar M. G. (2000) The regulator of G protein signaling family. Annu. Rev. Pharmacol. Toxicol. 40, 235–271 [DOI] [PubMed] [Google Scholar]
- 7. Hollinger S., Taylor J. B., Goldman E. H., Hepler J. R. (2001) RGS14 is a bifunctional regulator of Gαi/o activity that exists in multiple populations in brain. J. Neurochem. 79, 941–949 [DOI] [PubMed] [Google Scholar]
- 8. Evans P. R., Lee S. E., Smith Y., Hepler J. R. (2014) Postnatal developmental expression of regulator of G protein signaling 14 (RGS14) in the mouse brain. J. Comp. Neurol. 522, 186–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee S. E., Simons S. B., Heldt S. A., Zhao M., Schroeder J. P., Vellano C. P., Cowan D. P., Ramineni S., Yates C. K., Feng Y., Smith Y., Sweatt J. D., Weinshenker D., Ressler K. J., Dudek S. M., Hepler J. R. (2010) RGS14 is a natural suppressor of both synaptic plasticity in CA2 neurons and hippocampal-based learning and memory. Proc. Natl. Acad. Sci. U.S.A. 107, 16994–16998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vellano C. P., Lee S. E., Dudek S. M., Hepler J. R. (2011) RGS14 at the interface of hippocampal signaling and synaptic plasticity. Trends Pharmacol. Sci. 32, 666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takesono A., Cismowski M. J., Ribas C., Bernard M., Chung P., Hazard S., 3rd, Duzic E., Lanier S. M. (1999) Receptor-independent activators of heterotrimeric G-protein signaling pathways. J. Biol. Chem. 274, 33202–33205 [DOI] [PubMed] [Google Scholar]
- 12. De Vries L., Fischer T., Tronchère H., Brothers G. M., Strockbine B., Siderovski D. P., Farquhar M. G. (2000) Activator of G protein signaling 3 is a guanine dissociation inhibitor for Gαi subunits. Proc. Natl. Acad. Sci. U.S.A. 97, 14364–14369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shu F. J., Ramineni S., Amyot W., Hepler J. R. (2007) Selective interactions between Giα1 and Giα3 and the GoLoco/GPR domain of RGS14 influence its dynamic subcellular localization. Cell. Signal. 19, 163–176 [DOI] [PubMed] [Google Scholar]
- 14. Oner S. S., An N., Vural A., Breton B., Bouvier M., Blumer J. B., Lanier S. M. (2010) Regulation of the AGS3.Gαi signaling complex by a seven-transmembrane span receptor. J. Biol. Chem. 285, 33949–33958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oner S. S., Maher E. M., Breton B., Bouvier M., Blumer J. B. (2010) Receptor-regulated interaction of activator of G-protein signaling-4 and Gαi. J. Biol. Chem. 285, 20588–20594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Willard F. S., Willard M. D., Kimple A. J., Soundararajan M., Oestreich E. A., Li X., Sowa N. A., Kimple R. J., Doyle D. A., Der C. J., Zylka M. J., Snider W. D., Siderovski D. P. (2009) Regulator of G-protein signaling 14 (RGS14) is a selective H-Ras effector. PLoS One 4, e4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shu F. J., Ramineni S., Hepler J. R. (2010) RGS14 is a multifunctional scaffold that integrates G protein and Ras/Raf MAP kinase signalling pathways. Cell. Signal. 22, 366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cho H., Kozasa T., Takekoshi K., De Gunzburg J., Kehrl J. H. (2000) RGS14, a GTPase-activating protein for Giα, attenuates Giα- and G13α-mediated signaling pathways. Mol. Pharmacol. 58, 569–576 [DOI] [PubMed] [Google Scholar]
- 19. Traver S., Bidot C., Spassky N., Baltauss T., De Tand M. F., Thomas J. L., Zalc B., Janoueix-Lerosey I., Gunzburg J. D. (2000) RGS14 is a novel Rap effector that preferentially regulates the GTPase activity of Gαo. Biochem. J. 350, 19–29 [PMC free article] [PubMed] [Google Scholar]
- 20. Kimple R. J., De Vries L., Tronchère H., Behe C. I., Morris R. A., Gist Farquhar M., Siderovski D. P. (2001) RGS12 and RGS14 GoLoco motifs are Gαi interaction sites with guanine nucleotide dissociation inhibitor activity. J. Biol. Chem. 276, 29275–29281 [DOI] [PubMed] [Google Scholar]
- 21. Mittal V., Linder M. E. (2004) The RGS14 GoLoco domain discriminates among Gαi isoforms. J. Biol. Chem. 279, 46772–46778 [DOI] [PubMed] [Google Scholar]
- 22. Blumer J. B., Lanier S. M. (2014) Activators of G protein signaling exhibit broad functionality and define a distinct core signaling triad. Mol. Pharmacol. 85, 388–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abramow-Newerly M., Roy A. A., Nunn C., Chidiac P. (2006) RGS proteins have a signalling complex: interactions between RGS proteins and GPCRs, effectors, and auxiliary proteins. Cell. Signal. 18, 579–591 [DOI] [PubMed] [Google Scholar]
- 24. Vellano C. P., Maher E. M., Hepler J. R., Blumer J. B. (2011) G protein-coupled receptors and resistance to inhibitors of cholinesterase-8A (Ric-8A) both regulate the regulator of g protein signaling 14 RGS14·Gαi1 complex in live cells. J. Biol. Chem. 286, 38659–38669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vellano C. P., Shu F. J., Ramineni S., Yates C. K., Tall G. G., Hepler J. R. (2011) Activation of the regulator of G protein signaling 14-Gαi1-GDP signaling complex is regulated by resistance to inhibitors of cholinesterase-8A. Biochemistry 50, 752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vellano C. P., Brown N. E., Blumer J. B., Hepler J. R. (2013) Assembly and function of the regulator of G protein signaling 14 (RGS14)·H-Ras signaling complex in live cells are regulated by Gαi1 and Gαi-linked GPCRs. J. Biol. Chem. 288, 3620–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goswami D., Devarakonda S., Chalmers M. J., Pascal B. D., Spiegelman B. M., Griffin P. R. (2013) Time window expansion for HDX analysis of an intrinsically disordered protein. J. Am. Soc. Mass Spectrom. 24, 1584–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pascal B. D., Willis S., Lauer J. L., Landgraf R. R., West G. M., Marciano D., Novick S., Goswami D., Chalmers M. J., Griffin P. R. (2012) HDX workbench: software for the analysis of H/D exchange MS data. J. Am. Soc. Mass Spectrom. 23, 1512–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 30. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Berman D. M., Kozasa T., Gilman A. G. (1996) The GTPase-activating protein RGS4 stabilizes the transition state for nucleotide hydrolysis. J. Biol. Chem. 271, 27209–27212 [DOI] [PubMed] [Google Scholar]
- 33. Chen C. K., Wieland T., Simon M. I. (1996) RGS-r, a retinal specific RGS protein, binds an intermediate conformation of transducin and enhances recycling. Proc. Natl. Acad. Sci. U.S.A. 93, 12885–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Watson N., Linder M. E., Druey K. M., Kehrl J. H., Blumer K. J. (1996) RGS family members: GTPase-activating proteins for heterotrimeric G-protein α-subunits. Nature 383, 172–175 [DOI] [PubMed] [Google Scholar]
- 35. Goswami D., Callaway C., Pascal B. D., Kumar R., Edwards D. P., Griffin P. R. (2014) Influence of domain interactions on conformational mobility of the progesterone receptor detected by hydrogen/deuterium exchange mass spectrometry. Structure 22, 961–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Landgraf R. R., Goswami D., Rajamohan F., Harris M. S., Calabrese M. F., Hoth L. R., Magyar R., Pascal B. D., Chalmers M. J., Busby S. A., Kurumbail R. G., Griffin P. R. (2013) Activation of AMP-activated protein kinase revealed by hydrogen/deuterium exchange mass spectrometry. Structure 21, 1942–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marciano D. P., Dharmarajan V., Griffin P. R. (2014) HDX-MS guided drug discovery: small molecules and biopharmaceuticals. Curr. Opin. Struct. Biol. 28, 105–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mixon M. B., Lee E., Coleman D. E., Berghuis A. M., Gilman A. G., Sprang S. R. (1995) Tertiary and quaternary structural changes in Giα1 induced by GTP hydrolysis. Science 270, 954–960 [DOI] [PubMed] [Google Scholar]
- 39. Chung K. Y., Rasmussen S. G., Liu T., Li S., DeVree B. T., Chae P. S., Calinski D., Kobilka B. K., Woods V. L., Jr., Sunahara R. K. (2011) Conformational changes in the G protein Gs induced by the β2 adrenergic receptor. Nature 477, 611–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Soundararajan M., Willard F. S., Kimple A. J., Turnbull A. P., Ball L. J., Schoch G. A., Gileadi C., Fedorov O. Y., Dowler E. F., Higman V. A., Hutsell S. Q., Sundström M., Doyle D. A., Siderovski D. P. (2008) Structural diversity in the RGS domain and its interaction with heterotrimeric G protein α-subunits. Proc. Natl. Acad. Sci. U.S.A. 105, 6457–6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tesmer J. J. (2009) Structure and function of regulator of G protein signaling homology domains. Prog. Mol. Biol. Transl. Sci. 86, 75–113 [DOI] [PubMed] [Google Scholar]
- 42. Bosch D. E., Willard F. S., Ramanujam R., Kimple A. J., Willard M. D., Naqvi N. I., Siderovski D. P. (2012) A P-loop mutation in Galpha subunits prevents transition to the active state: implications for G-protein signaling in fungal pathogenesis. PLoS Pathog. 8, e1002553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Du Q., Macara I. G. (2004) Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell 119, 503–516 [DOI] [PubMed] [Google Scholar]
- 44. Bosch D. E., Kimple A. J., Sammond D. W., Muller R. E., Miley M. J., Machius M., Kuhlman B., Willard F. S., Siderovski D. P. (2011) Structural determinants of affinity enhancement between GoLoco motifs and G-protein α subunit mutants. J. Biol. Chem. 286, 3351–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kiel C., Wohlgemuth S., Rousseau F., Schymkowitz J., Ferkinghoff-Borg J., Wittinghofer F., Serrano L. (2005) Recognizing and defining true Ras binding domains II: in silico prediction based on homology modelling and energy calculations. J. Mol. Biol. 348, 759–775 [DOI] [PubMed] [Google Scholar]
- 46. Mittal V., Linder M. E. (2006) Biochemical characterization of RGS14: RGS14 activity towards G-protein α subunits is independent of its binding to Rap2A. Biochem. J. 394, 309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tu Y., Popov S., Slaughter C., Ross E. M. (1999) Palmitoylation of a conserved cysteine in the regulator of G protein signaling (RGS) domain modulates the GTPase-activating activity of RGS4 and RGS10. J. Biol. Chem. 274, 38260–38267 [DOI] [PubMed] [Google Scholar]
- 48. Ishii M., Fujita S., Yamada M., Hosaka Y., Kurachi Y. (2005) Phosphatidylinositol 3,4,5-trisphosphate and Ca2+/calmodulin competitively bind to the regulators of G-protein-signalling (RGS) domain of RGS4 and reciprocally regulate its action. Biochem. J. 385, 65–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Popov S. G., Krishna U. M., Falck J. R., Wilkie T. M. (2000) Ca2+/calmodulin reverses phosphatidylinositol 3,4,5-trisphosphate-dependent inhibition of regulators of G protein-signaling GTPase-activating protein activity. J. Biol. Chem. 275, 18962–18968 [DOI] [PubMed] [Google Scholar]
- 50. Evans P. R., Hepler J. R. (2012) Regulator of G protein signaling 14 (RGS14) interacts with calmodulin (CaM) in a calcium-dependent manner. 42nd Annual Meetiing of the Society for Neuroscience, New Orleans, October 13–17, 2012, Society for Neuroscience [Google Scholar]
- 51. Panja D., Bramham C. R. (2014) BDNF mechanisms in late LTP formation: a synthesis and breakdown. Neuropharmacology 76, 664–676 [DOI] [PubMed] [Google Scholar]
- 52. Kennedy M. B., Beale H. C., Carlisle H. J., Washburn L. R. (2005) Integration of biochemical signalling in spines. Nat. Rev. Neurosci. 6, 423–434 [DOI] [PubMed] [Google Scholar]
- 53. Hollinger S., Ramineni S., Hepler J. R. (2003) Phosphorylation of RGS14 by protein kinase A potentiates its activity toward Gαi. Biochemistry 42, 811–819 [DOI] [PubMed] [Google Scholar]
- 54. Hollinger S., Hepler J. R. (2004) Methods for measuring RGS protein phosphorylation by G protein-regulated kinases. Methods Mol. Biol. 237, 205–219 [DOI] [PubMed] [Google Scholar]
- 55. Traver S., Splingard A., Gaudriault G., Gunzburg J. D. (2004) The RGS (regulator of G protein signalling) and GoLoco domains of RGS14 co-operate to regulate Gi-mediated signalling. Biochem. J. 379, 627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]