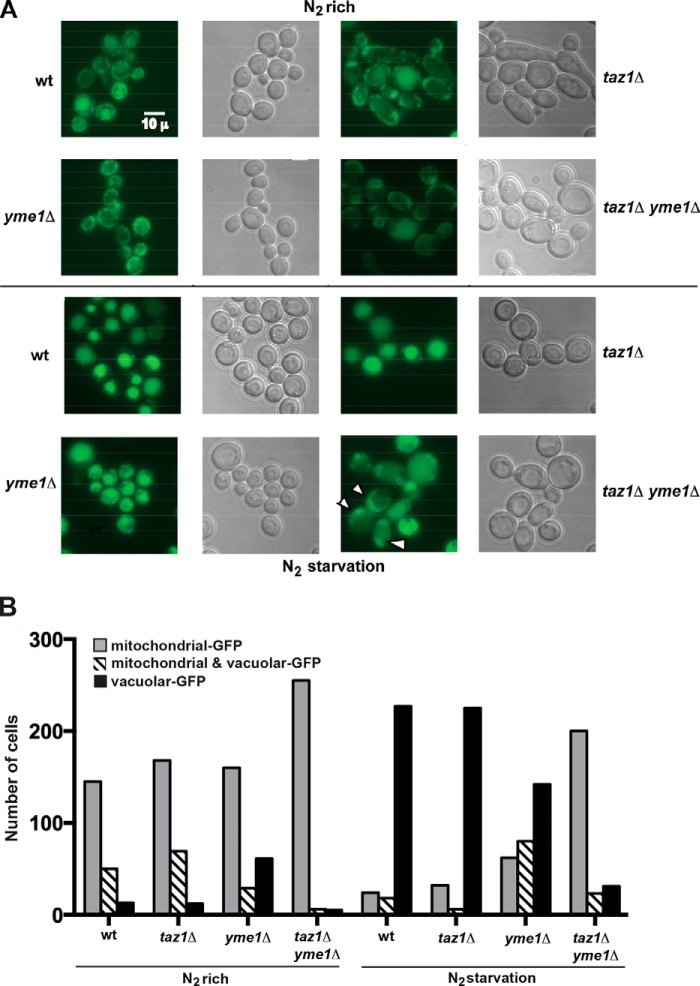
FIGURE 6.
Mitophagy is inefficient in taz1Δ yme1Δ yeast. The Om45 allele was tagged with a GFP epitope in all of the indicated strains, and these cells were grown in YPL medium to 0.8–1.5 A600. The cells were then washed twice in distilled water, and an aliquot of cells was saved and processed for microscopy and Western blotting prior to resuspension and growth in SD−N medium to induce N2 starvation. After 24 h of growth in SD−N medium, the cells were harvested and processed for fluorescence microscopy. A, representative fluorescence and differential interference contrast microscopy, indicating the cellular localization of Om45-GFP before and after 24 h of nitrogen starvation. After nitrogen starvation, Om45-GFP was predominantly localized to the vacuole (an indication of mitophagy) in the majority of the cells of all of the yeast strains except for taz1Δyme1Δ yeast (arrows indicate cells with GFP signal outside the vacuole). B, the number of GFP-positive mitochondria and vacuoles from 250 cells were determined from three experiments for each strain under each growth condition. The S.E. was less than 5% for each experiment.