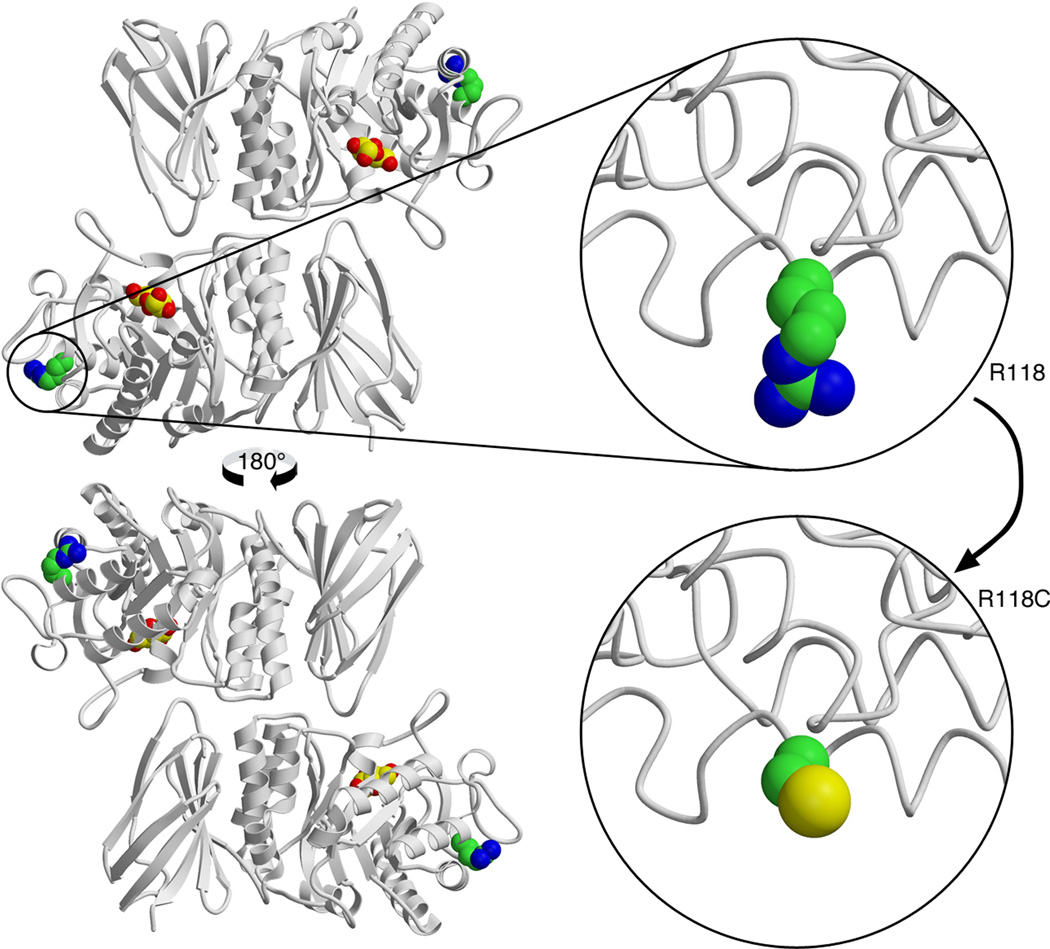
Figure 1.
Molecular structure of the wild-type mature human α-galactosidase A enzyme and modeling of the p.(Arg118Cys) variant: the change to cysteine is easily accommodated in the three-dimensional structure of the enzyme because the arginine is a surface residue and there is plenty of room to substitute the cysteine side chain. However, the cysteine side chain has different chemistry, which can interfere with the correct folding of the disulfide bonds required for the structure, or it could interfere with the binding of other molecules – like the chaperones BiP (binding immunoglobulin protein) and calnexin –, that are required for the folding and trafficking of the α-galactosidase A the lysosome. The structural prediction is that the protein should be active when it folds, but the efficiency of folding and trafficking will be reduced. This is consistent with the results of in vitro overexpression experiments [28].