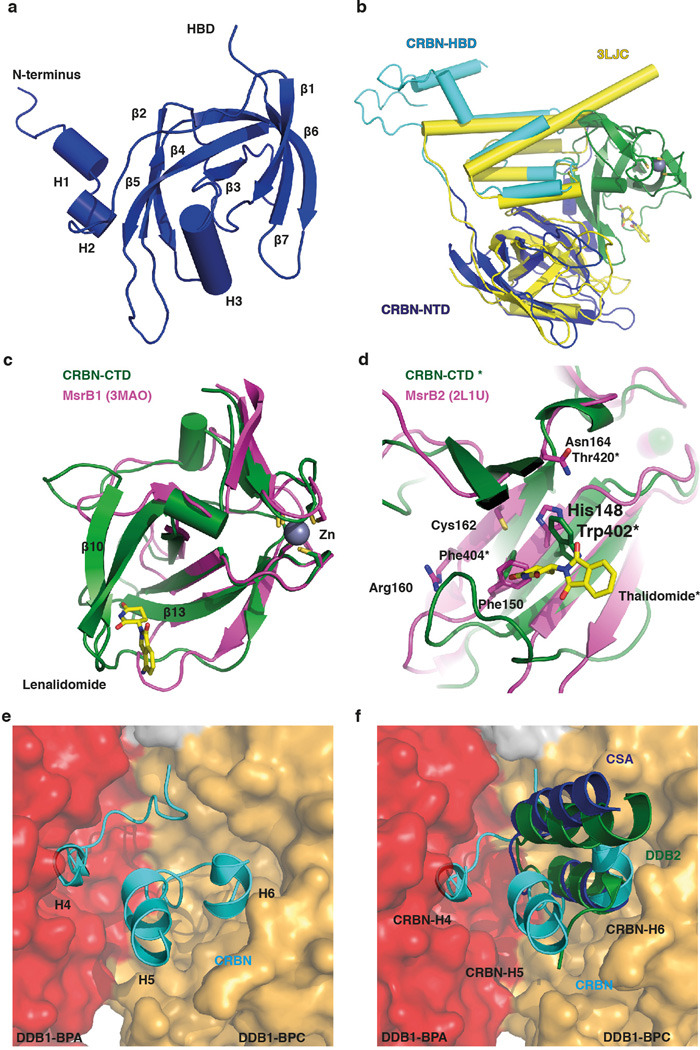
Extended Data Figure 2. Structure of the DDB1-CRBN complex bound to thalidomide and derivatives.
(a) Close-up view and topology of the ggCRBN N-terminal domain (NTD). (b) Overlay of the ggCRBN-NTD (dark-blue) and the helical bundle domain (HBD) (light-blue) with the Lon-protease domain coloured yellow (PDB: 3LJC - RMSD of 2.7 Å over 178 residues aligned). How Lon-proteases recognise their substrates is unclear at present. As both the NTD and the HBD are present in the Lon-fold, it is conceivable that CRBN originated from a fusion of a Lon-protease with a PUA fold-containing enzyme (see below). In the course of divergent evolution, the helical element of the Lon-fold appeared to be utilised for DDB1 binding. (c) The ggCRBN C-terminal domain (CTD: residues aa318-aa427) harbours the thalidomide, lenalidomide, or pomalidomide binding pocket and displays structural similarity to pseudouridine synthase and archaeosine transglycosylase (PUA) folds. The alignment with the methionine sulfoxide reductase subfamily, the closest structural ggCRBN orthologue (PDB: 3MAO - RMSD of 2.0 Å over 79 residues) is shown. The PUA domain fold family frequently carries a conserved metal coordination site, which in ggCRBN appears to bind Zn2+ and is coordinated by Cys325, 328, 393 and 396. (d) Residues involved in the catalytic activity in the MsrB2 reductase (Trp103, His148, Phe150, Arg160, Cys162 and Arg162). The catalytic cysteine (Cys 162) residue is not conserved in ggCRBN, which makes it unlikely that ggCRBN acts as an MsrB2-like reductase. Interestingly, however, residues equivalent to Trp402 and Phe404 involved in ggCRBN thalidomide binding are also involved in substrate binding and catalysis in MsrB2. The conserved binding patch surrounding lenalidomide may thus act as a substrate binding interface, with thalidomide possibly blocking binding of a hitherto unknown protein substrate, or a specific post-translation modification. (e) The ggCRBN interaction with DDB1 is mediated through the HBD motif utilizing ggCRBN α-helices H5: 224–233, H6: 244–249, and H4: 200–204, a 310 helix. ggCRBN helices H5 and H6 together with the intervening loop segment form extensive interactions with the DDB1-BPC propeller, with additional contributions from residues ggCRBN 190–195. ggCRBN H4 and the preceding loop (197–205) form interactions with DDB1-BPA. ggCRBN binding to BPA occurs through a H1 310 helix located at the central cavity of the WD40 propeller, the narrow side of the WD40-propeller cone where ligands typically bind. (f) ggCRBN binding to DDB1 differs from that previously seen in DCAF-DDB1 complexes, where the majority of interactions outside the WD40 propeller domain originated from a consecutive helix-loop-helix (HLH) motif that predominantly contacts DDB1-BPC (golden-brown surface). Extensive interactions between a DCAF and DDB1-BPA domain (red surface) have not been observed previously, in either the DDB1 co-crystal structures with DDB2 (pdb: 3EI3; green), CSA (pdb: 4A11; dark blue) or other HLH motifs to which DDB1 binds. The novel DDB1 binding mode found in CRBN precluded prior sequence-based identification and suggested considerable plasticity of DCAF binding to DDB1.