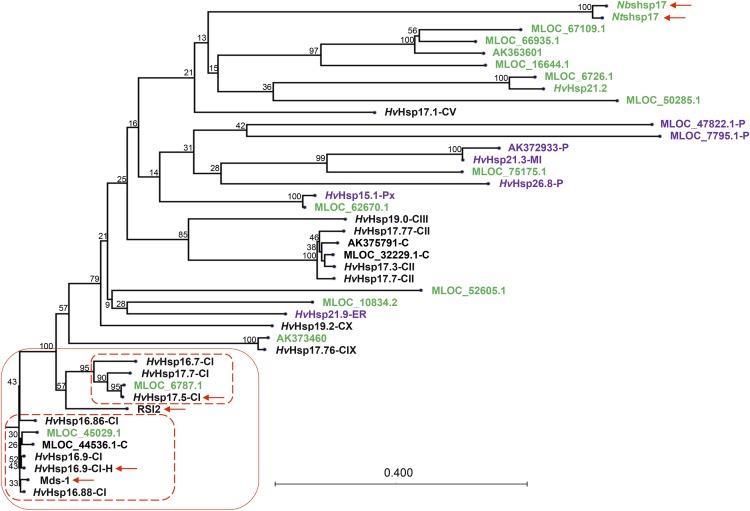
Figure 4.
Amino acid-based phylogenetic tree of all 37 identified full-length barley sHsps, tomato RSI2, Ntshsp17, and Nbshsp17. The lower left branches mainly contain cytosolic (black) sHsps with high similarity to each other. Those with noncytosolic and unknown localizations are shown in purple and green, respectively. Letters following the molecular weight indicate their localizations: C, cytosolic; ER, endoplasmic reticulum; MI, mitochondria; P, chloroplast; and Px, peroxisome. The two sHsps identified in the Y2H screen, RSI2, Nthsp17, Nbhsp17, and Mds1 are indicated by arrows. The naming follows Reddy et al. (2014) for those starting with HvHsp. The MLOC names are from the annotation of the barley genome. There are four sequences solely based on cDNA clones, with GenBank accession numbers given.