Abstract
The ligands of the Bone Morphogenetic Protein (BMP) family of developmental signaling molecules are often under the control of complex cis-regulatory modules and play diverse roles in vertebrate development and evolution. Here, we investigated the cis-regulatory control of stickleback Bmp6. We identified a 190 bp enhancer ~2.5 kilobases 5’ of the Bmp6 gene that recapitulates expression in developing teeth and fins, with a core 72 bp sequence that is sufficient for both domains. By testing orthologous enhancers with varying degrees of sequence conservation from outgroup teleosts in transgenic reporter gene assays in sticklebacks and zebrafish, we found that the function of this regulatory element appears to have been conserved for over 250 million years of teleost evolution. We show that a predicted binding site for the TGFβ effector Smad3 in this enhancer is required for enhancer function and that pharmacological inhibition of TGFβ signaling abolishes enhancer activity and severely reduces endogenous Bmp6 expression. Finally, we used TALENs to disrupt the enhancer in vivo and find that Bmp6 expression is dramatically reduced in teeth and fins, suggesting this enhancer is necessary for expression of the Bmp6 locus. This work identifies a relatively short regulatory sequence that is required for expression in multiple tissues and, combined with previous work, suggests that shared regulatory networks control limb and tooth development.
Keywords: Bone Morphogenetic Protein, enhancer, tooth development, stickleback, zebrafish, Bmp6, TGFβ
Introduction
Bone Morphogenetic Protein (BMP) ligands, the largest subfamily of TGFβ ligands, play multiple essential roles during vertebrate development (Hogan, 1996; Kingsley, 1994; Massagué, 2012), including during craniofacial and tooth development (Nie et al., 2006). Many vertebrate organs develop through reciprocal permissive and instructive signaling between adjacent epithelial and mesenchymal tissues, often involving multiple BMP ligands (Bellusci et al., 1996; Dassule and McMahon, 1998; Dudley et al., 1999; Jung et al., 1998). These pleiotropic functions of BMP ligands are orchestrated by typically large, modular, regulatory regions, which work together to drive complex spatiotemporally restricted expression patterns (Pregizer and Mortlock, 2009).
In humans, regulatory variation in Bmp genes has been associated with developmental disorders including brachydactyly and other birth defects (Dathe et al., 2009; Justice et al., 2012), as well as colorectal cancer (Houlston et al., 2008; Lubbe et al., 2012). In other animals, variation in the expression of Bmp genes has also been associated with major evolved changes in morphology, including beak shape in Darwin’s finches (Abzhanov et al., 2004), jaw size and shape in cichlid fish (Albertson et al., 2005), and tooth number in stickleback fish (Cleves et al 2014).
While the cis-regulatory architecture of Bmp2, Bmp4, Bmp5, and Bmp7 has been studied in mice (Adams et al., 2007; Chandler et al., 2007; Guenther et al., 2008; Jumlongras et al., 2012), less is known about Bmp6 and Bmp gene regulation in other vertebrates. Although not required for viability in the mouse, Bmp6 is required for axial skeletal patterning (Solloway et al., 1998), kidney function (Dendooven et al., 2011), and physiological iron regulation (Andriopoulos et al., 2009). Non-coding variants in human Bmp6 have been associated with human height variation (Gudbjartsson et al., 2008; Wood et al., 2014), as well as orofacial clefting birth defects (Shi et al., 2012). A cis-regulatory allele of stickleback Bmp6 with reduced Bmp6 expression in developing tooth tissue has recently been shown to be associated with evolved increases in tooth number in derived freshwater sticklebacks, likely adaptive for the shift in diet in freshwater sticklebacks relative to their marine ancestors (Cleves et al., 2014).
BMP signaling plays complex and, in general, poorly understood roles during the development of placodes. During tooth development, multiple Bmp genes are expressed dynamically in developing odontogenic epithelia and mesenchyme (Aberg et al., 1997; Vainio et al., 1993). Several lines of evidence reveal BMP signaling plays activating roles during odontogenesis. First, epithelial BMP4 activates Msx expression in the mesenchyme, and exogenous BMP from a bead (Bei and Maas, 1998; Chen et al., 1996) or transgene (Zhao et al., 2000) can partially rescue tooth development in Msx1 mutant mice. Second, in mice, teeth arrest at the bud-to-cap transition in Bmpr1a mutants (Andl et al., 2004; Liu et al., 2005). Third, exogenous BMP4 beads can induce molar development in mice (Kavanagh et al., 2007). Fourth, in fish, pharmacological inhibition of BMP signaling can inhibit tooth formation in cichlids (Fraser et al., 2013). In contrast, other evidence supports BMP signaling playing inhibitory effects during the development of teeth and other placodes. In mice, Pax9 expression marks early dental mesenchyme, and BMP2 and BMP4 inhibit Pax9 expression (Neubüser et al., 1997). In zebrafish, inhibition of BMP signaling produces supernumerary teeth with altered morphology (Jackman et al., 2013). During development of both feather and hair placodes, BMPs play inhibitory roles (Botchkarev et al., 1999; Jung et al., 1998; Mou et al., 2006, 2011), and suppression of epithelial BMP signaling is required for hair placode induction (reviewed in Biggs and Mikkola, 2014). Together these results suggest that complex positive and negative interactions between epithelial and mesenchymal BMPs are critical for placode development, yet the regulation of these interactions remains less well understood.
Despite the major role BMP signaling plays during tooth development, little is known about the cis-regulatory sequences that drive dynamic Bmp expression in early developing odontogenic epithelia and mesenchyme. In mice, a late-stage ameloblast enhancer has been identified for the Bmp4 gene (Feng et al., 2002); however this enhancer is not reported to be active during embryogenesis, or in dental mesenchyme. A second enhancer of mouse Bmp4 has been described that is active during embryogenesis and drives expression in dental epithelium but not mesenchyme (Jumlongras et al., 2012). Tooth epithelial and mesenchymal enhancers of the mouse Bmp2 gene have been localized to a ~150 kb region 3’ of Bmp2 (Chandler et al., 2007), however these enhancers have not yet been further mapped, and in general, cis-regulation of BMPs in dental mesenchyme is poorly understood. Furthermore, since mice are monophyodonts that form one wave of primary teeth and no replacements, less is known about cis-regulatory elements that drive expression in developing and replacement teeth in polyphyodont vertebrates (such as fish) that replace their teeth continuously. Because of the recently identified cis-regulatory allele of Bmp6 associated with evolved changes in stickleback tooth number (Cleves et al., 2014) and to dissect epithelial and mesenchymal cis-regulation of vertebrate BMP signaling, we sought to begin to identify the cis-regulatory architecture of the stickleback Bmp6 gene.
Methods
Animal statement and fish husbandry
All animal work was approved by the Institutional Animal Care and Use Committee of the University of California-Berkeley (protocol number R330). Sticklebacks (Gasterosteus aculeatus) were raised in ~10% seawater (3.5 g/l Instant Ocean salt, 0.217 ml/l 10% sodium bicarbonate) at 18° C, and crosses were generated by in vitro fertilization. Zebrafish (Danio rerio) were raised in a recirculating system under standard conditions, and embryos were collected either from natural spawning or in vitro fertilization and raised at 28.5 degrees (Westerfield, 2007).
BAC Isolation and Recombineering
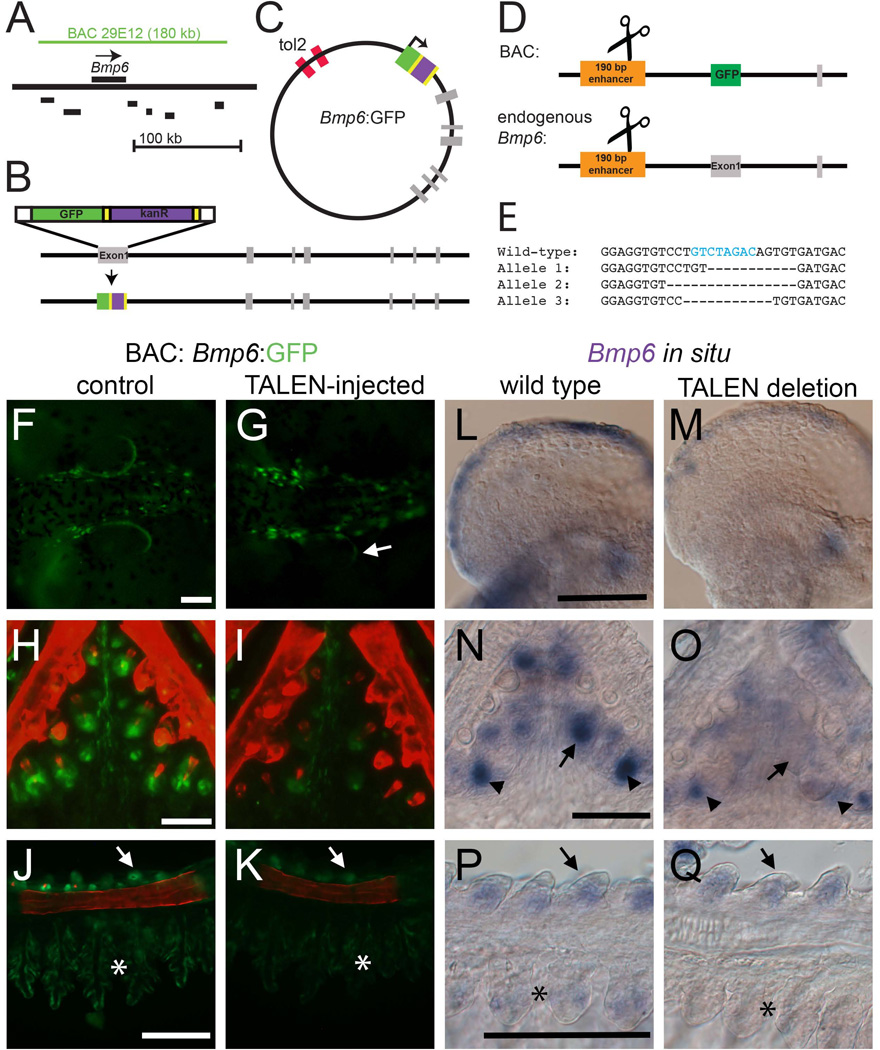
Bacterial Artificial Chromosomes (BACs) from the CHORI-213 and CHORI-215 (Salmon River marine and Paxton benthic freshwater stickleback, respectively) BAC libraries were identified by overgo screening (Ross et al., 1999) using the following overgoes: 5’-TGTGACGTTGACCTCAGCTAGACT-3’ and 5’-GAGGATTTAAACCGGGAGTCTAGC −3’. BAC ends were sequenced using Sp6 and T7 primers and mapped to the stickleback genome using the UCSC browser. BAC CHORI-215-29E12 was chosen for reporter analysis because Bmp6 was relatively centrally located in the BAC. Inverted Tol2 sites were recombineered into the Lox511 site of the pTarbac2.1 backbone according to Suster et al. (2011) using primers PTARBAC_tol2FWD and PTARBAC_tol2REV, and ampicillin resistance was used to select successfully recombineered BAC clones. To place GFP into exon 1 of Bmp6 as a reporter, a GFP/kanamycin resistant cassette was amplified from pGFP-FRT-Kan-FRT (Suster et al., 2011) using primers GFP_Bmp6_for and GFP_Bmp6_rev (Table S1), which contained 50 bp homology to the beginning and end of the first exon of stickleback Bmp6, respectively. This construct was then recombineered into the BAC containing iTol2 sites to produce the final reporter BAC (see Fig. 6A–C).
Fig. 6. The 5’ 190 bp enhancer is necessary for Bmp6 expression.
(A) Schematic of the genomic location of the 180 kb CHORI BAC29E12 with respect to Bmp6 and nearby genes (coding regions shown in black are Ipo4, Pdcd6, Txndc5, Muted, Eef1e1, and Slc35b3 from left to right). (B) Recombineering strategy for introducing GFP into the first exon of Bmp6; grey bars indicate exons. (C) Final circular BAC with inverted Tol2 sites for transposition and GFP reporter (not to scale). (D) Strategy for introducing TALEN lesions into the 190 bp 5’ enhancer. The same TALENs were used to target the enhancer in stable transgenic BAC fish and at the endogenous Bmp6 locus (diagram not to scale). (E) Sequences of stable mutant enhancer alleles. For the endogenous locus targeting, F2 fish trans-heterozygous for two different enhancer mutations were generated. Fish in (M) carried alleles 1 and 2; fish in (O) and (Q) carried alleles 1 and 3. The predicted Smad3 binding site is indicated with blue text in the wild type sequence. (F, G) In the reporter BAC, TALEN injection frequently severely reduced GFP expression from the pectoral fin relative to controls at 5 dpf. A small patch of mosaic, unaffected GFP is indicated with the arrow in (G). (H, I) TALEN injection also eliminated much of the Bmp6 tooth expression (I). (J, K) GFP expression was also reduced in gills (asterisk) and slightly reduced in the gill rakers (arrowhead). (L–M). Mutations in the enhancer caused a reduction in pectoral fin expression relative to wild-type siblings. (N, O) Bmp6 expression was also lost in tooth epithelia (arrows), but was not entirely lost in mesenchyme (arrowheads). (P, Q) Expression was also noticeably reduced in gills (asterisk), though gill raker expression (arrows) appears similar to wild-type sibling controls. Scale bars = 100 µm.
Enhancer Constructs
The vector for the stickleback 2.8 kb enhancer/promoter construct was generated using pENTRbasGFP and pTolDest (Villefranc et al., 2007) using Gateway cloning to produce a construct with the carp β-actin basal promoter (Scheer and Campos-Ortega, 1999) upstream of EGFP, flanked by Tol2 sites (Urasaki et al., 2006). Next, a 2,810 bp sequence upstream of the predicted Bmp6 transcriptional start site was PCR amplified from BAC CHORI-213-256N24 using primers Gac_3kb_for and Gac_3kb_rev and cloned upstream of the carp β-actin promoter using a ClaI restriction site. Blocks of conserved sequences within the 2.8 kb construct were identified as CS1, CS2, and CS3 from the UCSC 8 species Multiz conservation track (see Fig. 1A). These sequences were cloned into ClaI site of the carp β-actin reporter construct using primers shown in Table S1. CS1 was cloned with Gac_3kb_for and Gac_CS1_rev. CS2 was cloned with Gac_CS2_for and Gac_CS2_rev. CS3 was cloned with Gac_CS3_for and Gac_3kb_rev. CS2+3 was cloned with Gac_CS2_for and Gac_3kb_rev. Because the CS1 fragment drove weak expression with the β-actin promoter, we switched to using a well-characterized zebrafish hsp70 promoter construct, which we found to drive much brighter expression in transgenic stickleback embryos. CS1 and CS2+3 were also cloned into the hsp70 promoter construct for additional testing using the same genomic primer sequences but with Nhe and BamHI restriction sites in place of ClaI. The 190 bp and 72 bp enhancer sequences were amplified from the 2.8 kb construct with primers indicated in Table S1 and cloned into the hsp70 construct.
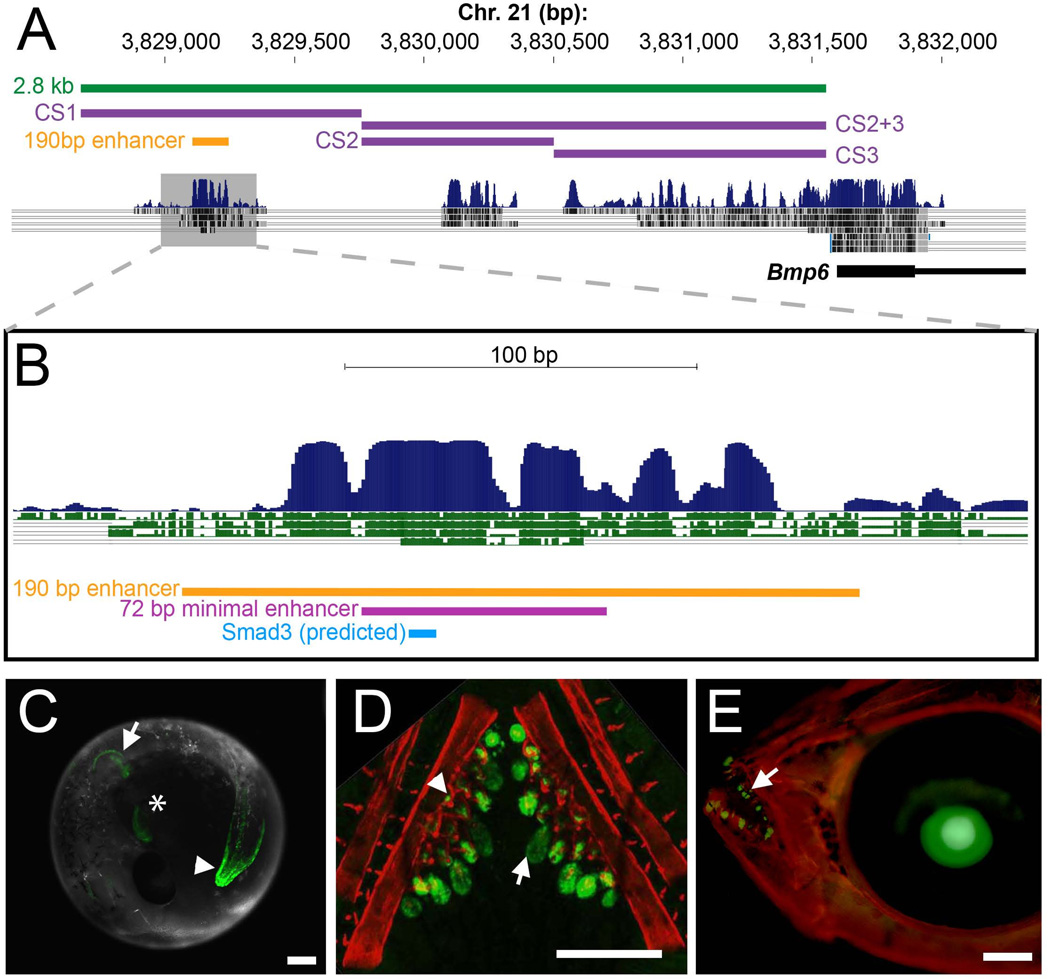
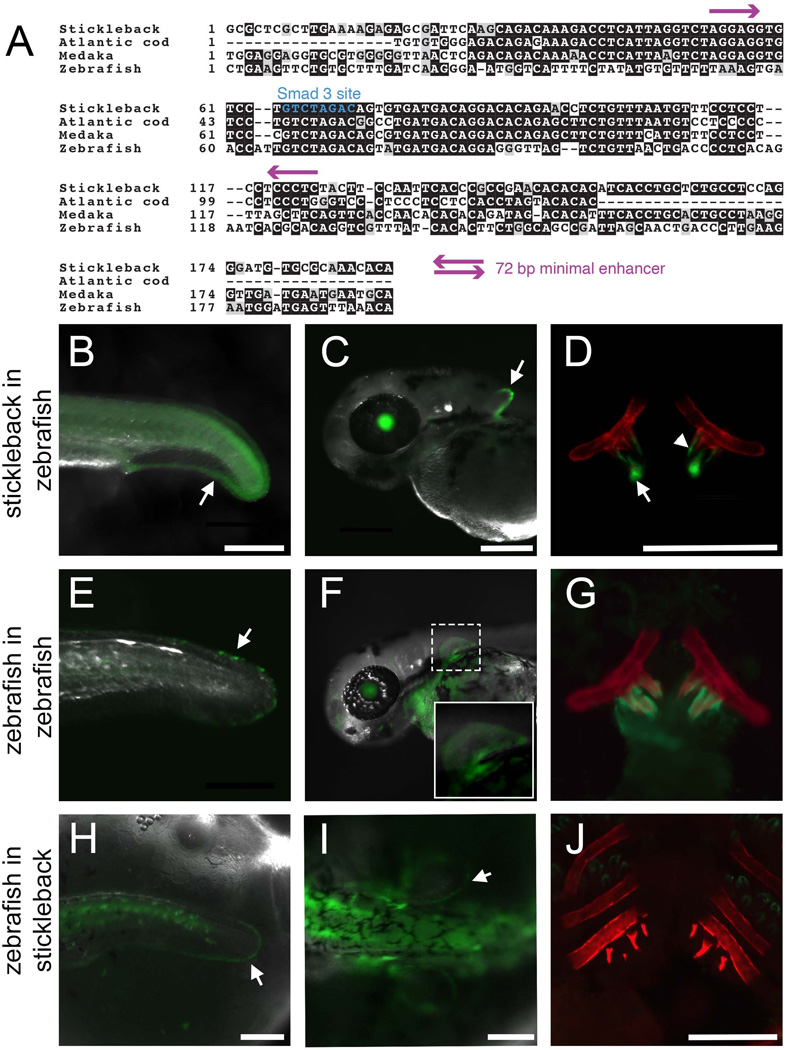
Fig. 1. A conserved 190 bp enhancer upstream of Bmp6 drives gene expression in several domains.
(A) The 5’ region of stickleback Bmp6 from the UCSC genome browser (http://genome.ucsc.edu/). The region of genomic DNA used in the 2.8 kb enhancer construct (see Fig. S3) is shown in green, conserved sequences (CS) 1–3 are shown in purple, and the subcloned 190 bp enhancer is shown in yellow. The first exon and part of the first intron of Bmp6 are shown in thick and thin black lines, respectively (bottom). Conservation peaks and alignments (dark blue and grey) are shown from the 8-Species MultiZ track. (B) Zoom in on the middle of CS1, approximately 2.5 kb upstream of the predicted Bmp6 transcription start site. The 190 bp enhancer, the 72 bp minimal enhancer (see Fig. S6), and a predicted Smad3 binding site (see Fig. 3–4) are shown in yellow, pink, and blue, respectively. The conservation track is shown as dark blue peaks, above green alignments showing conservation to medaka, tetraodon, fugu, and zebrafish, from top to bottom. (C) GFP reporter expression pattern driven by the 190 bp enhancer in a 5 dpf (stage 22, (Swarup, 1958)) stickleback embryo. Strong expression was seen in the distal edge of the developing pectoral fin (arrow), the heart (asterisk), and the distal edge of the median fin (arrowhead). (D) Confocal projection of GFP reporter expression in the ventral pharyngeal tooth plate in a ~10 mm stickleback fry. Expression was observed in the epithelium of developing tooth germs (arrow) and the odontogenic mesenchyme (arrowhead) in the cores of ossified teeth. Bones are fluorescently stained with Alizarin red. (E) GFP reporter expression in the oral teeth (arrow) of a 30 dpf stickleback fry. GFP in the lens is an internal control for the zebrafish hsp70 promoter used in the transgenic construct. Scale bars = 200 µm.
The orthologous enhancer sequences were identified in other teleost genomes using the UCSC genome browser (genome.ucsc.edu) to identify sequence conservation. Zebrafish and medaka (Oryzias latipes) wild-type genomic DNA was isolated by standard phenol-chlorofom extraction and enhancers were amplified using primers (Table S1) designed from the respective genome assemblies (zv9/danRer7 and oryLat2) and cloned into the hsp70 promoter construct. The Atlantic cod (Gadus morhua) enhancer DNA sequence was identified by sequence conservation on contig CAEA01327401 of the Atlantic cod genome assembly (UCSC, gadMor1). This short, unassembled contig is flanked by repetitive sequence, but the intervening sequence contains a 94 bp stretch that has 92.4% sequence identity to the stickleback enhancer and is likely the orthologous sequence. We synthesized a 130 bp construct of Atlantic cod sequence by using two primers for amplification (Gmo_for and Gmo_rev, see Table S1) and two additional overlapping oligonucleotides as template (Gmo_temp1 and Gmo_temp2). The template oligonucleotides were added to standard Phusion (NEB) PCR reaction at a concentration of 0.05 µM to amplify the full 130bp sequence, which was then cloned into the Tol2 construct as described above.
Sequence Analysis
Sequence alignments were generated using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) (Larkin et al., 2007) and Boxshade (http://www.ch.embnet.org/software/BOX_form.html). Binding sites were predicted with the UniProbe database (http://the_brain.bwh.harvard.edu/uniprobe/) (Newburger and Bulyk, 2009) and PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) (Farre et al., 2003; Messeguer et al., 2002).
Imaging and Microscopy
Transgenic lines were imaged using a Leica DM2500 compound microscope equipped with a Leica DFC500 camera, a Leica M165FC dissecting microscope equipped with a DFC340 FX camera, or a Zeiss 700 confocal microscope. Transgenic fish were fixed for 4 hours at 4°C in either 4% paraformaldehyde in 1X PBS or 10% neutral buffered formalin. For Alizarin red fluorescent counterstaining of GFP lines, 0.01% Alizarin red was added to the fixative. Tooth number was counted on the DM2500 with TX2 filter to visualize Alizarin-stained teeth. Tooth germs with GFP+ epithelia were counted on photographs of GFP fluorescence.
Fish injections and line generation
Transposase mRNA was produced from the pCS2-TP plasmid (Kawakami et al., 2004) with the mMessage mMachine SP6 in vitro transcription kit (Ambion) according to manufacturer’s instructions and purified with a Qiagen RNeasy column. Zebrafish injections were performed with 25 ng/µL plasmid DNA and 37.5 ng/µL transposase and 0.05% phenol red as previously described (Fisher et al., 2006). Because stickleback embryos are much larger than zebrafish embryos, the DNA and RNA concentrations were increased to 37.5 and 75 ng/µL respectively. Stable transgenic lines were generated by outcrossing injected fish to non-transgenic fish and visually screening for fluorescent transgenic offspring. At least two stable lines were observed for each construct to ensure fluorescent patterns were due to the transgene and not artifacts of the transgene integration sites.
Site directed mutagenesis
Mutagenesis primers were designed using the online Quickchange tool (http://www.genomics.agilent.com/primerDesignProgram). For constructs containing multiple mutations, the mutagenesis was performed in multiple rounds. Mutagenesis reactions were performed with 125 ng of each primer, 50 ng plasmid template, 200 µM dNTPs, and Pfu Turbo polymerase and buffer. Cycling conditions were 95°C for 30 seconds, followed by 16 cycles of 95°C / 30 s, 55°C / 60 s, and 68°C /780 s. Primer sequences can be found in supplementary Table 1; the mutated sequences are shown in Fig. 3A. DpnI was added immediately after cycling, and the reaction was incubated for 1 hr at 37°C, then immediately transformed into Top10 chemically competent E. coli cells.
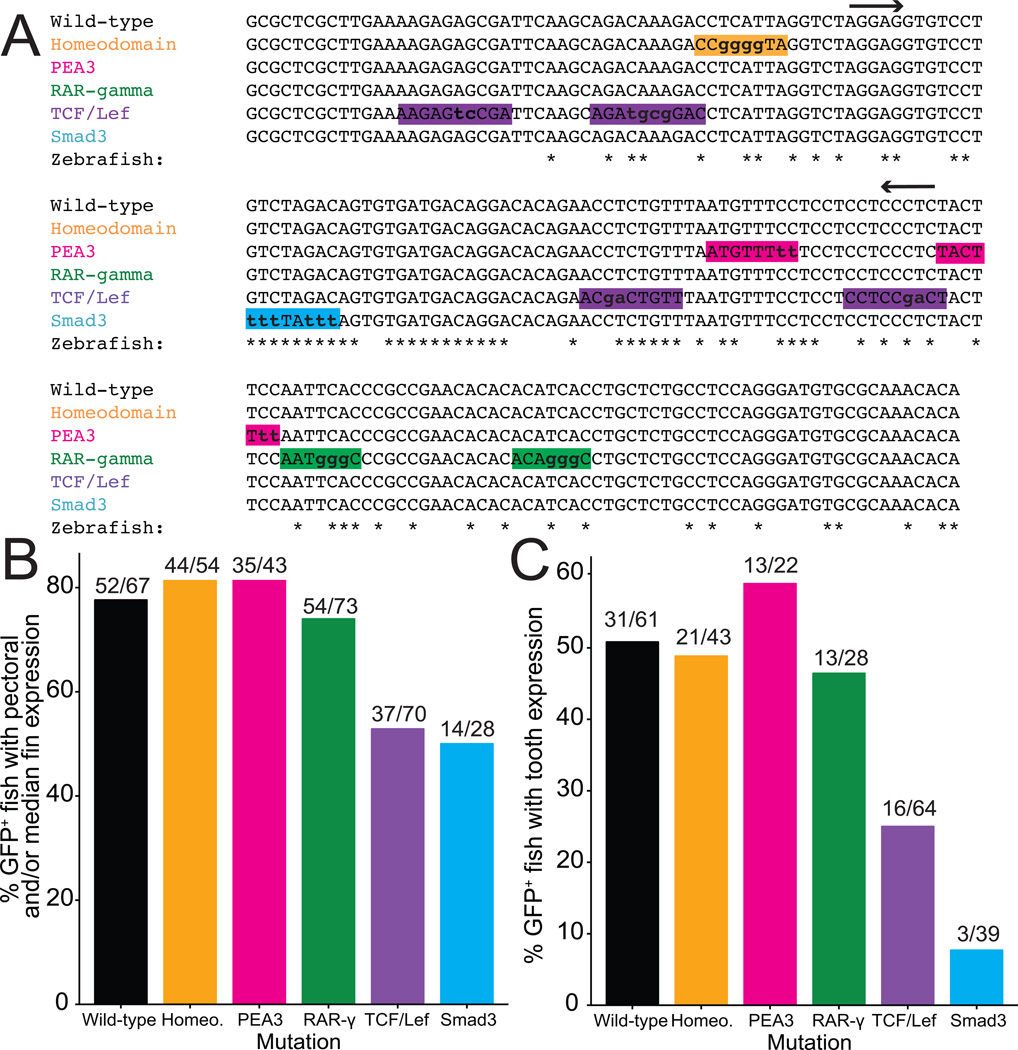
Fig. 3. Mutations in a predicted Smad3 binding site severely reduce enhancer function.
(A) Binding sites predicted by UniProbe and PROMO are highlighted with a unique color for each signaling pathway. Highlighted sequences represent the “predicted sequence” from PROMO or the “K-mer” from UniProbe. Mutated base pairs are shown with lowercase letters. Nucleotide positions conserved to zebrafish are indicated with an asterisk, and arrows indicate the 72 bp minimal enhancer sequence. (B–C) Sticklebacks were injected with each mutated construct and scored for pectoral fin and/or median fin expression at 5 dpf (B) and oral and/or pharyngeal tooth expression at 12–13 dpf (C). Frequency of expression in these domains is shown as a percentage of the total number of GFP-positive fish (scored as GFP expression driven by the hsp70 promoter anywhere at 5 dpf or in the lens at 12–13 dpf) on the y-axis.
Drug treatments
SB431542 and XAV939 (Sigma) were dissolved in DMSO to concentrations of 100 µM and 10 µM, respectively. The drug was then diluted into stickleback water or zebrafish system water to working concentrations (25–100 µM for SB431542 and 5–10 µM for XAV939). A DMSO vehicle control was done in parallel with all drug treatments. Drug treatment was performed in 6- or 24-well cell culture dishes. Sticklebacks were treated from 2 dpf to 5 dpf for observation of pectoral and median fin expression, and for 5–7 days post-hatching for observation of tooth GFP. Zebrafish were treated beginning at 10 hpf for observation of median fin and beginning at 24 hpf for pectoral fin and tooth expression. For multiday treatments, fresh solution was applied every 48 hours until the end of the experiment.
In situ hybridization (ISH)
Bmp6 in situ hybridization was performed on embryos and newly-hatched juveniles as previously described (Cleves et al., 2014). For pharyngeal tooth and gill in situs, the branchial skeleton was dissected out of the embryo and cut along the dorsal midline prior to the hybridization step.
Mutagenesis using TALENs
TAL Effector Nucleases (TALENs) were targeted to the predicted Smad3 binding site within the 190 bp enhancer using TAL Effector Nuclear Targeter 2.0 (https://talent.cac.cornell.edu/) using the Cermak architecture (Cermak et al., 2011; Doyle et al., 2012). TALEN plasmids were generated using the RVDs shown in Table S4. TALEN mRNAs were produced with the Mmessage Mmachine kit (Ambion), purified with Qiagen RNeasy columns, and injected into one-cell stickleback embryos at a concentration of 40 ng/µL for each mRNA plus 0.05% phenol red. Embryos and juvenile fish were screened for lesions in the Smad3 site by screening for loss of an XbaI cut site in a 144 bp PCR product amplified with primers Gac_190_for and Gac_72_rev (see Fig. 4G). F1 animals with deletions visible on a 2% agarose gel (~15 bp or larger) were crossed to generate animals used in in situ hybridization. Because the F1 parents carried different TALEN-induced lesions, the F2 animals used for ISH were transheterozygotes for two slightly different alleles of the enhancer deletion (see Fig. 6E).
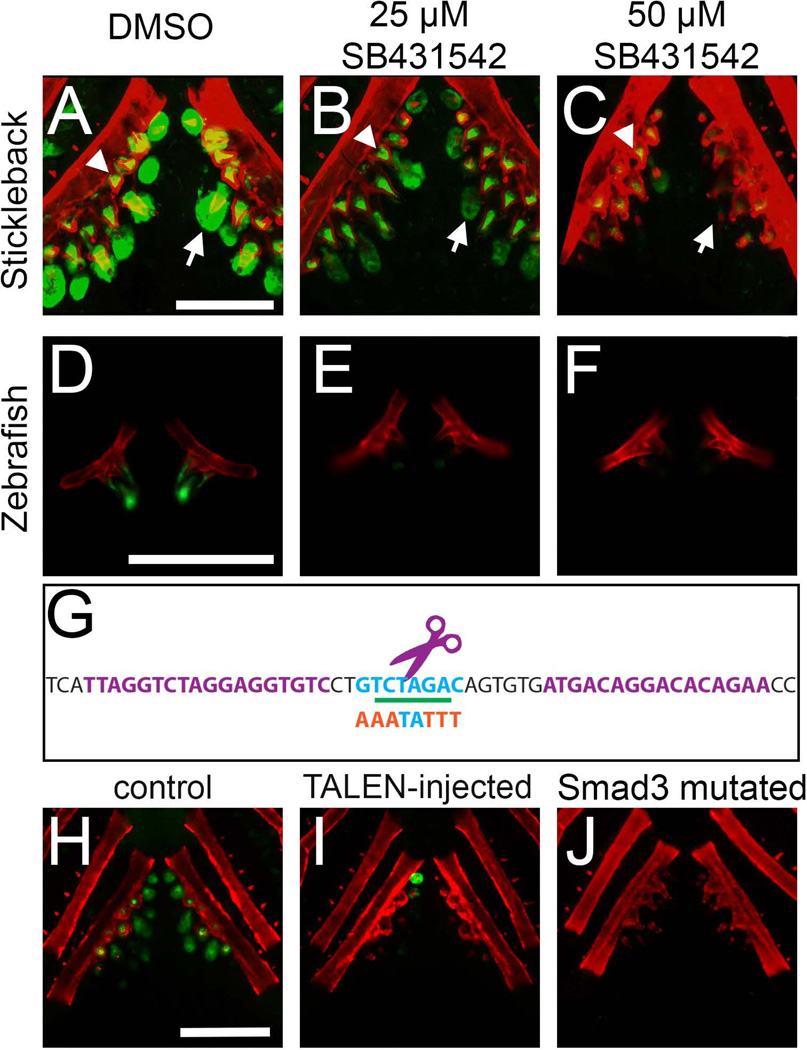
Fig. 4. Pharmacological disruption of TGFβ signaling or TALEN-induced mutations of the predicted Smad3 binding site reduce enhancer activity.
(A–C) Treatment of stickleback fry for 7 days in SB431542 (an ALK5 inhibitor) severely reduced GFP expression driven by the 190 bp enhancer in a dose-dependent manner. Expression was severely reduced in the epithelia (arrows), but not mesenchyme (asterisks), of pharyngeal teeth at both low (25 µM, B) and high (50 µM, C) doses relative to controls (A). (D–F) SB431542 also eliminated GFP driven by the stickleback enhancer in a zebrafish trans environment. (G) The sequence targeted by TALENs contains a predicted Smad3 homodimer binding site (blue). The TALEN binding sites are indicated in purple text and the purple scissors indicate the approximate site of endonuclease activity. The XbaI site used for molecular screening is underlined in green, and the mutagenized sequence of the Smad3 binding site, indicated by orange letters, is shown below. (H–I) Injection of the TALENs into stable transgenic fish carrying the 190 bp reporter construct resulted in near complete loss of GFP expression in 95% of injected animals (I) relative to controls (H). Residual GFP seen in (I) is likely the result of the mosaicism of TALEN-induced lesions. (J). Mutating the predicted Smad3 binding site resulted in a loss of GFP expression in both epithelium and mesenchyme of pharyngeal teeth in 3/3 stickleback lines observed. Bone is fluorescently counterstained with Alizarin red. Scale bars = 200 µm.
Results
A Bmp6 reporter BAC recapitulates endogenous Bmp6 expression
To begin to identify the cis-regulatory architecture of the stickleback Bmp6 gene, we generated a Bmp6 reporter line by identifying a bacterial artificial chromosome (CHORI BAC215-29E12) containing 180 kb of sequence starting ~52 kb upstream of Bmp6. Inverted Tol2 sequences were recombineered into the backbone of this BAC, and the first exon of Bmp6 was replaced with GFP coding sequence. This transgenic construct drove GFP reporter expression in a variety of tissues throughout development (Fig. S1), including the embryonic tailbud following somitogenesis (3.5 dpf), the embryonic heart and ventrolateral cells in the pharyngeal region (4 dpf), the distal edge of the developing pectoral fin, and the distal edge of the median fin (5 dpf). After hatching (10–15 dpf), expression was seen in oral and pharyngeal teeth, the pericardium, cells surrounding the opercle and branchiostegal rays, gill buds, and gill rakers.
We compared this transgene expression pattern to the expression pattern of endogenous Bmp6 mRNA via in situ hybridization. We observed Bmp6 expression in nearly all of the same domains as the reporter BAC (Fig. S2), including the tailbud (at 3.5 dpf), heart, the distal edges of the median and pectoral fins (at 5 dpf), gills, gill rakers, and in the previously described (Cleves et al., 2014) epithelium and mesenchyme of developing teeth (assayed at ~12 dpf). However, several domains observed by in situ hybridization were not observed in the BAC transgenic line, including the notochord, the dorsal medial diencephalon, the eyes, and the ears (Fig. S2), suggesting that regulatory elements lying outside of the 180 kb of genomic sequence contained within the BAC control these Bmp6 expression domain.
A conserved 190 bp enhancer drives tooth, median fin, and pectoral fin expression in both stickleback and zebrafish
To begin to identify regulatory elements contained within this 180 kb genomic interval containing Bmp6, we first cloned a construct containing ~2.8 kb of sequence immediately upstream of stickleback Bmp6 containing regions of sequence conserved among other teleosts (Fig. 1A). This construct drove GFP expression in a number of tissues that were similar to expression patterns driven by the BAC (Fig. S3, compare to Fig. S1), including the tailbud, the heart, pectoral and median fins, oral and pharyngeal teeth, gills, and the pericardium. Other domains driven by the BAC were not observed in the 5’ construct, including gill rakers, opercle, and branchiostegal rays; these domains are likely driven by more distal regulatory elements contained within the BAC but excluded from the 2.8 kb sequence. Combined, these results suggest that much of the regulatory information for Bmp6 is contained within the 2.8 kb upstream sequence, but that other regulatory elements drive additional expression domains.
We hypothesized that the different anatomical sites of expression driven by the 2.8 kb fragment result from multiple anatomically specific enhancers within this sequence. We first tested three non-overlapping subclones, each containing a block of evolutionarily conserved sequence (Fig. 1A). While the most 5’ subclone (CS1) drove robust reporter gene expression in most domains of the 2.8 kb fragment, neither the middle (CS2) nor 3’ subclone (CS3) drove detectable GFP expression in fins, teeth, or other domains driven by the 2.8 kb fragment at the 3–5 dpf or post-hatching (10–13 dpf) stages. Furthermore, a construct containing CS2 + CS3 also drove no detectable pattern of GFP with either the β-actin or hsp70 promoter. Next, we focused on the 5’-most region (CS1), and tested a 190 bp fragment highly conserved within teleosts (Fig. 1B). This 190 bp fragment drove robust GFP expression in the distal edges of the pectoral and median fins, and oral and pharyngeal teeth (Fig. 1C–E). Within developing teeth, GFP expression was observed in the inner dental epithelium (IDE) for all constructs (Fig. S4) as well as the interior mesenchyme of mature functional teeth (Fig. 1D), similar to endogenous Bmp6 expression during tooth development (Cleves et al., 2014). Robust tooth GFP expression was seen in all teeth at all stages examined including in juveniles and adults, suggesting tooth enhancer activity is present in both primary and replacement teeth (Fig. 1D–E, data not shown). Some domains, including the gills, were lost when CS1 was reduced to the 190 bp fragment, suggesting that flanking sequence is required for these domains. When the orientation of the enhancer was flipped with respect to the hsp70 promoter, 77% (38/49) of injected fish had pectoral and/or median fin expression at 5 dpf, and 69% (27/39) had oral and/or pharyngeal tooth expression at 13 dpf. This result suggests that this enhancer functions regardless of orientation to the promoter. Combined, our results suggest that most domains driven by the 2.8 kb enhancer are driven by the short 190 bp conserved sequence. This 190 bp minimal sequence does not differ between marine and freshwater sticklebacks, though several marine-freshwater sequence differences exist in the surrounding sequences of CS1.
Conservation of cis regulatory elements and trans machinery in teleosts
Because we used evolutionary sequence conservation to identify the 190 bp minimal enhancer and the sequence was partially conserved to zebrafish, we hypothesized that this 190 bp stickleback enhancer would show similar activity in transgenic zebrafish. Stickleback and zebrafish are ~250 million years divergent (Near et al., 2012) and share only 3 short blocks (totaling 28 bp, Fig. 2A) of perfectly conserved nucleotides in the middle of the enhancer. However, the stickleback enhancer robustly drove a highly similar expression pattern in zebrafish, with expression in the distal edges of the median and pectoral fins, and pharyngeal tooth epithelium and mesenchyme (Fig. 2B–D), suggesting that the trans factors activating the enhancer are conserved in distantly related teleosts. We next asked whether the orthologous sequence from the zebrafish genome had similar enhancer activity in both zebrafish and sticklebacks. A construct containing 477 bp of sequence from the orthologous region of the zebrafish genome drove weak expression in these expression domains (distal edges of median and pectoral fins, and teeth) in a subset of transgenic zebrafish offspring obtained (Fig. 2E–G and Table S2). In sticklebacks, seven stable transgenic lines with the zebrafish sequence driving GFP had no fin expression, although one transgenic line displayed very faint expression in the distal edges of the median and pectoral fins (Fig. 2H–I). None of the eight lines had GFP expression in teeth (Fig. 2J). Therefore, sticklebacks and zebrafish likely share the trans machinery sufficient to drive expression from the stickleback sequence, but the cis regulatory information present in the zebrafish orthologous sequence is not sufficient to drive tooth expression in the stickleback trans environment.
Fig. 2. Evolutionary functional conservation of the Bmp6 enhancer in teleosts.
(A) Sequence alignments of four teleost sequences relative to the 190 bp stickleback enhancer. The perfectly conserved Smad3 dimer binding site is marked in blue, and purple arrows mark the boundaries of the 72 bp minimal enhancer (see Fig. S6). (B–D) The stickleback sequence reporter construct stably integrated into the zebrafish genome drove expression in the distal edge of the median fin at 24 hpf (arrow in B), the distal edge of the pectoral fin at 48 hpf (arrow in C), and tooth epithelium (arrow) and mesenchyme (arrowhead) at 5 dpf (D). (E–G) A 477 bp construct of zebrafish genomic sequence centered around the conserved sequence of the enhancer drove similar, but weaker expression in the median fin of a 33 hpf zebrafish (arrow in E), pectoral fins of a 48 hpf zebrafish (inset of F), and teeth of a 5 dpf zebrafish (G). (H–I) Although not detected in seven of eight stable lines, in one of eight stable lines, the zebrafish sequence drove faint expression in the distal edges of the median fin (arrow in H) and pectoral fins (arrow in I) of 5 dpf stickleback. However, no expression was detected in tooth germs in newly hatched fry in any line (J). See Table S2 for quantification of expression domains of transgenic lines. Bone is fluorescently stained with Alizarin red in (D, G, J). Scale bars = 200 µm.
Because the zebrafish enhancer shows much less sequence conservation to sticklebacks relative to other teleosts (Fig. 2A), we hypothesized that the loss of robustness and loss of tooth expression may be unique to the zebrafish cis-regulatory element. We generated constructs containing the orthologous enhancer sequences of a beloniform (medaka) and a gadiform (Atlantic cod), which fall between zebrafish and sticklebacks in the teleost phylogeny (Near et al., 2012). We found that sequences from both additional species drove expression in fins and teeth in both stickleback and zebrafish embryos (Fig. S5, Table S2), although the cod enhancer appeared to be slightly less robust (Table S2).
Based on the apparent partial conservation of enhancer function in zebrafish and the conserved activities of the medaka and cod enhancers, we further shortened the stickleback enhancer to contain the sequence most highly conserved among teleosts, a 72 bp sequence near the center of the 190 bp construct, and hypothesized that it would drive the tooth, median fin, and pectoral fin expression domains. In support of this hypothesis, this construct in a stable line of zebrafish was sufficient to drive strong GFP expression in teeth and median and pectoral fins (Fig. S6). Notably, the heart domain driven by this construct was considerably brighter relative to the 190 bp enhancer, suggesting that this short sequence may have lost additional repressor elements that limit expression in the heart. A similar pattern of brighter heart expression was observed in stickleback injected with this construct compared to the 190 bp larger construct (data not shown). These results suggest that the flanking conserved sequences are not required for the basic enhancer pattern in fins and teeth, but may be important for fine-tuning the transcriptional output.
A predicted Smad3 binding site is required for enhancer function
To identify candidate transcription factor binding sites within the 190 bp enhancer, we used UniProbe and PROMO (Newburger and Bulyk, 2009; Farre et al., 2003; Messeguer et al., 2002) and found predicted binding sites of transcription factors in several signaling pathways involved in developmental regulation: FGF (PEA3), retinoic acid (RAR-γ), Wnt (TCF/Lef), and TGFβ (Smad3), as well as a predicted homeodomain binding site (Fig. 3 A). We were particularly interested in the homeodomain binding site given the known crosstalk between the Msx1 and Bmp4 genes during mouse tooth development (Bei and Maas, 1998; Chen et al., 1996; Jumlongras et al., 2012), and the predicted TCF/Lef sites, given the previously described roles of Wnt signaling regulating Bmp4 dental mesenchyme expression in mice (Fujimori et al., 2010; O’Connell et al., 2012). We quantified the number of stickleback embryos showing pectoral and/or median fin, as well as pharyngeal and/or oral tooth expression, when injected with constructs containing mutated binding sites. The mutation of TCF/Lef and Smad3 binding sites significantly decreased the percentage of fish with median and/or pectoral fin expression domains, whereas the predicted PEA3, RAR-γ, and homeodomain mutations did not (Fig. 3B). Likewise, only the mutations in predicted TCF/Lef and Smad3 sites affected tooth expression, with especially reduced expression when the predicted Smad3 binding site was mutated (Fig. 3C). We made stable zebrafish lines for each of the Smad3 and TCF/Lef mutated enhancers and found that the Smad3-mutated reporter construct did not drive robust expression in zebrafish fins or teeth, while the TCF/Lef mutated construct did drive these domains, albeit at apparently reduced levels (Fig. S7). Since the Smad3-mutated construct did not drive fin or tooth expression in zebrafish, we generated a stable line in sticklebacks and found that this line similarly did not drive detectable median fin, pectoral fin, or tooth expression (Fig. 4J). Therefore, the predicted Smad3 site is required for normal enhancer output, while TCF/Lef sites may be responsible for expression level but not tissue specificity.
A small molecule inhibitor of TGFβ signaling, but not a small molecule inhibitor of Wnt signaling, abolishes enhancer function
Since the predicted Smad3 binding site was necessary for enhancer function, we hypothesized that reducing TGFβ signaling (mediated by Smad3) would result in a loss of expression driven by the enhancer. To pharmacologically inhibit TGFβ signaling, we treated transgenic sticklebacks and zebrafish embryos with SB431542, a specific inhibitor of ALK4/5 phosphatase activity that abrogates TGF-β signaling in zebrafish (Inman et al., 2002; Sun et al., 2006). After 6 days of treatment in sticklebacks, GFP expression driven by the 190 bp enhancer was reduced in a dose-dependent manner in the epithelium, but not mesenchyme, of developing pharyngeal teeth, with tooth epithelial expression abolished at 50 µM and reduced at 25 µM (Fig. 4A–C). Tooth mesenchymal expression was slightly diminished at 50 µM and apparently unaffected at 25 µM. Similarly, GFP reporter expression was lost in the pharyngeal teeth of newly hatched zebrafish upon treatment with SB431542 from 24 hpf until 5 dpf (Fig. 4D–F). In sticklebacks, we also saw a reduction, but not complete loss, of pectoral and median fin expression driven by the transgene upon treatment with SB431542 (Fig. S8), while the reduction was more severe in the fins of zebrafish. Combined with our site-directed mutagenesis of the Smad3 binding site result, these pharmacological data suggest that TGFβ signaling mediated by ALK4/5 (likely signaling via Smad3 binding) is necessary for tooth epithelium enhancer activity. However other signals likely contribute to the expression in the pectoral and median fins and tooth mesenchyme, as drug treatment did not completely abolish these expression domains in sticklebacks.
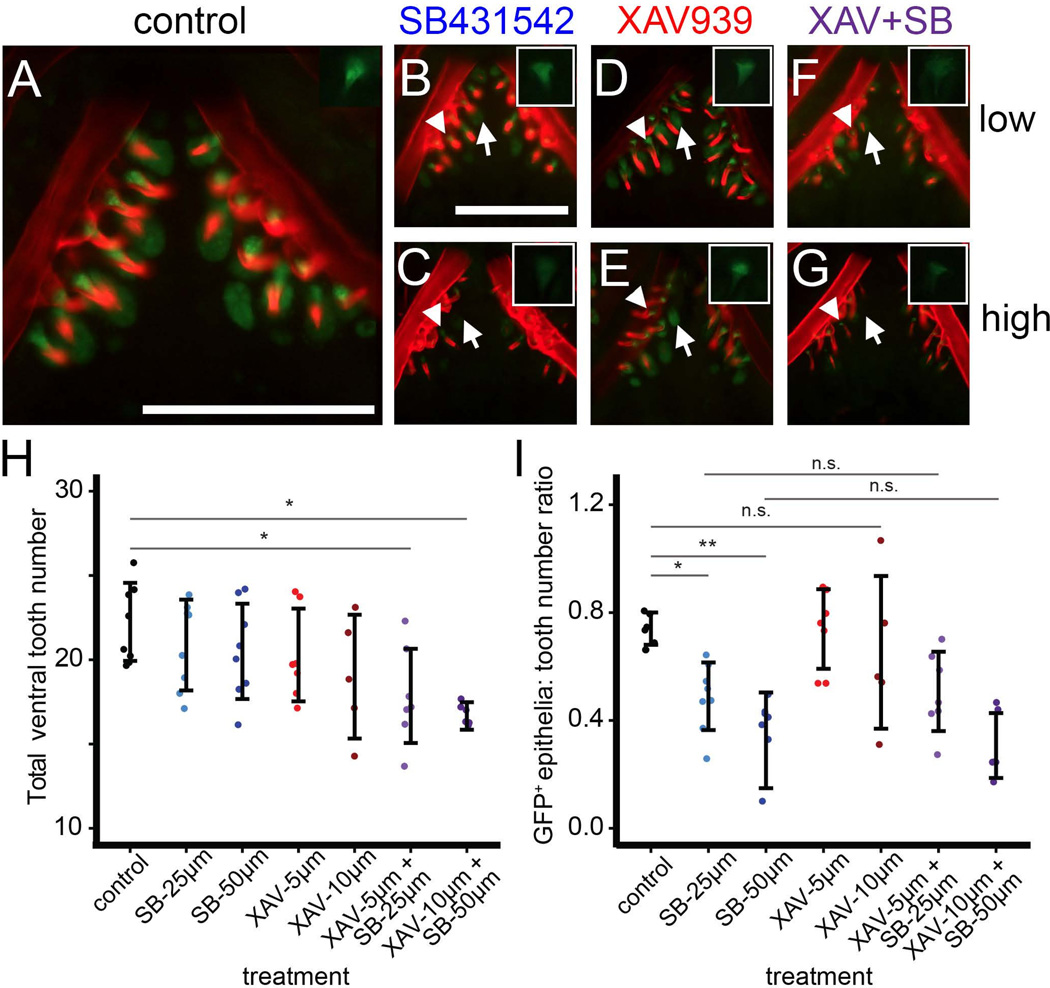
Since the mutation of TCF/Lef binding sites appeared to decrease enhancer activity in sticklebacks and zebrafish (Fig. 3, Fig. S7), we hypothesized that Wnt signaling might be an additional input into the 190 bp Bmp6 enhancer. To test this hypothesis, we treated transgenic fish with SB431542, XAV939 (a specific inhibitor of the Wnt signaling pathway that is active in zebrafish (Huang et al., 2009)), or both drugs in combination at low and high doses. Treatment with a high-dose combination of XAV939 and SB431542 decreased the standard length of fish (data not shown), possibly indicating a slight developmental delay. With XAV939 or SB431542 treatment alone, there was no effect of the drug on tooth number, suggesting that neither drug alone arrests tooth development. However, the two drugs in combination significantly reduced ventral pharyngeal tooth number (Fig. 5H), including at the low dose that did not affect fish standard length, suggesting that XAV939 is bioactive in sticklebacks and that reducing Wnt and TGFβ signaling together disrupts tooth development.
Fig. 5. Wnt signaling is not required for enhancer function, but Wnt and TGFβ are required for tooth development.
Newly hatched stickleback fry were treated with DMSO (control, A), SB431542 (B–C), XAV939 (D–E), or a combination of the two drugs at low (25µM for SB431542 and 5 µM for XAV939, F) or high (50 µM for SB431542 or 10 µM XAV939, G) doses for 5 days. Main panels show Alizarin red and GFP for the ventral tooth plate; insets show GFP only for mesenchyme of a single tooth from the dorsal tooth plate. (B, C) SB431542 reduced GFP in tooth epithelia (arrows) relative to control (A, and see Fig. 3). However, mesenchymal GFP (arrowhead, inset) was less severely reduced. (D, E) XAV939 alone did not affect GFP expression in epithelia (arrows) or mesenchyme (arrowheads) at either dose. (F, G) No strong additional effect on GFP expression was seen when XAV939 and SB431542 were combined, though mesenchymal GFP appeared slightly lower in the combined dose. (H) A combination of SB431542 and XAV939 significantly reduced ventral pharyngeal tooth number. (I) Treatment with SB431542, but not XAV939, decreased the number of green tooth epithelia relative to total ventral teeth (ratio is expressed as a decimal). XAV939 had no additional effect on green epithelia in combination with SB431542. Tukey HSD P-values of relevant comparisons are shown above with asterisks (*=P<0.05, ** =P<0.0005, n.s.=P>0.05). Scale bars = 200 µm
There was no obvious qualitatively detectable effect of XAV939 treatment on the intensity of enhancer expression in the teeth, either alone or in combination with SB431542 (Fig. 5; compare D and E to A, and compare F and G to B and C). However, tooth mesenchymal GFP in the combined drug treatment appeared slightly lower than with SB431542 treatment alone (insets of Fig. 5). Importantly, we never saw a complete loss of mesenchymal GFP with any drug treatment, but frequently saw complete loss of epithelial GFP with SB431542 treatment. To quantify the effect of drug treatment on epithelial GFP expression, we counted the number of GFP+ tooth epithelia (regardless of fluorescent intensity) in each treatment and expressed it as a ratio to the total number of Alizarin red-stained teeth. XAV939 had no effect on the relative number of GFP+ epithelia, while SB431542 had a strong, dose-dependent effect (Fig. 5I). In combination with SB431542, there was no additional effect of XAV939 on reporter expression (GFP+ epithelia in the combination treatments did not differ from treatment with SB431542 alone). Combined, our results suggest that SB431542, but not XAV939, affects enhancer activity and that simultaneous inhibition of Wnt and TGFβ signaling affects tooth development.
The 190 bp enhancer is necessary for Bmp6 expression
As an additional test of the importance of the predicted Smad3 binding site, we generated a pair of TALENs designed to induce mutations in this region of the enhancer (see Fig. 4G). This pair of TALENs was highly efficient at producing lesions, detected molecularly by loss of an XbaI restriction site, and confirmed by Sanger sequencing in a subset of individuals (Table S3; example deletions shown in Fig. 6E). Upon injection of these TALENs into a stable transgenic line of the 190 bp enhancer driving GFP, 95% of animals (40 of 42) showed partial or full loss of GFP fluorescence in the pectoral fins and median fin expression at 5 dpf. In those same animals, 95% of animals (39 of 41) also showed partial or complete loss of oral and/or pharyngeal tooth expression at 12–13 dpf (see example in Fig. 4I). Thus, the lesions generated by these TALENs are highly effective at disrupting activity driven by this 190 bp enhancer.
We next tested whether the sequence targeted by the TALENs was necessary for Bmp6 expression by injecting the TALENs into a stable transgenic line of the Bmp6:GFP BAC reporter. 91% (61/67) of animals had a reduction or complete loss of pectoral and median fin expression, and 89% (8/9) of dissected tooth plates showed severe reductions of GFP expression in the pharyngeal teeth (representative animals shown in Fig. 6 F–K). Notably, GFP expression in the embryonic and juvenile heart was detectable at seemingly unaffected levels in all animals, suggesting that the enhancer is not necessary for this expression domain. Additionally, gill expression appeared to be reduced but not completely eliminated in all animals observed (n=6), and gill raker expression was only slightly reduced. These data suggest the enhancer is required for some (e.g. pectoral fin, median fin, tooth epithelium), but not all domains of Bmp6 expression.
Next, we tested the role of the enhancer in driving endogenous Bmp6 expression by performing in situ hybridization for Bmp6 in fish trans-heterozygous for different TALEN-induced mutations in the predicted Smad3 binding site (Fig. 6E). In these trans-heterozygous fish, expression of Bmp6 was dramatically reduced in fins, tooth epithelia and gills, but gill raker expression appeared similar to wild-type controls (Fig. 6L–Q). Despite the severe loss of Bmp6 expression in tooth epithelia in mutant fish, expression in the mesenchyme of developing teeth was still detectable, although at apparently reduced levels (Fig. 6N–O). Thus, this enhancer is required to maintain normal levels of Bmp6 expression in developing fins and tooth epithelia.
TGFβ signaling is necessary for normal Bmp6 expression levels
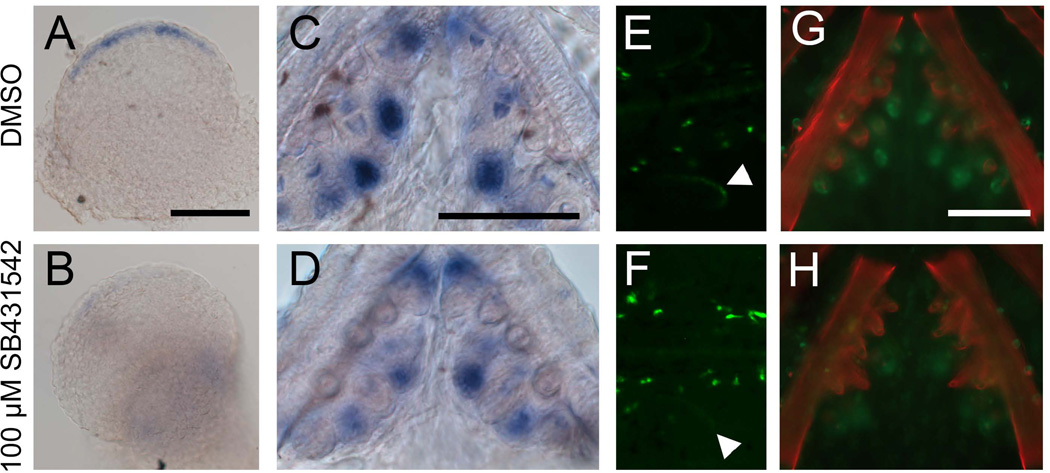
Since enhancer activity was lost upon treatment with a TGFβ inhibitor, and the enhancer is required for normal Bmp6 expression, we predicted that endogenous Bmp6 expression would likewise be reduced upon inhibition of TGFβ signaling. By in situ hybridization, pectoral fin and tooth epithelium expression of Bmp6 were both reduced upon 100 µM SB431542 treatment (Fig. 7A–D). SB431542 treatment also reduced GFP expression in reporter BAC animals in fins and teeth (Fig. 7E–H). The effect of the drug on BAC-driven GFP was not robustly observed with a 50 µM treatment (data not shown), despite the strong effect that this dose had on enhancer expression (Fig. 4). Together these data support a model in which TGFβ signaling is required for Bmp6 expression in teeth and fins and exerts its effect through the putative Smad3 binding site that is necessary for enhancer function.
Fig. 7. Treatment with TGFβ inhibitor SB431542 reduces Bmp6 expression.
(A–D) Sticklebacks were treated with 100 µM SB431542 or vehicle control from 2 to 5 dpf or for 5 days post-hatching, and Bmp6 expression was assayed by in situ hybridization. Drug treatment severely reduced Bmp6 expression in fins (A, B) and also reduced Bmp6 expression in tooth epithelia (C, D). Likewise, GFP driven by the Bmp6 locus in the reporter BAC was also reduced in fins (arrowheads in E, F) and teeth (G, H) after SB431542 treatment. Scale bars = 100 µm.
Discussion
A short, conserved enhancer with pleiotropic expression domains required for Bmp6 tooth and fin expression
Here we have identified a 190 base pair enhancer that is highly conserved in teleosts and is both necessary and sufficient for tooth and fin expression of stickleback Bmp6. Site-directed mutagenesis of a predicted Smad3 binding site and pharmacological experiments suggest this enhancer is TGFβ-responsive. Though this enhancer drives expression in several of Bmp6’s endogenous domains, our results suggest that like other Bmp genes, stickleback Bmp6 contains a complex cis-regulatory architecture composed of multiple modules driving expression in different domains. We detected embryonic expression domains of Bmp6 by in situ hybridization, such as the eye, ear, diencephalon, and notochord, that were not observed in the BAC reporter line, suggesting that the regulatory elements controlling these domains lie outside of the 180 kb of stickleback DNA included in the BAC. Furthermore, while TALEN mutations severely reduced expression in the fins and teeth, every BAC reporter fish injected with TALENs had GFP expression in the heart, suggesting that the enhancer is not required for heart expression. Thus, the short enhancer presented here contributes to a subset of the endogenous Bmp6 expression domains, with other domains likely driven by other enhancers greater than ~100 kb away. Evidence for long range distant enhancers of stickleback Bmp6 is expected, given the frequent finding of long distance regulatory elements for developmental regulatory genes, including other vertebrate Bmp genes (reviewed in Preziger and Mortlock, 2009). Interestingly, despite the presence of redundant “shadow” enhancers found in m any genes (Calle-Mustienes et al., 2005; Marinić et al., 2013; Perry et al., 2010), this enhancer appears to be required for several Bmp6 expression domains; additional enhancers did not appear to sufficiently compensate in driving Bmp6 expression when the 5’ enhancer was targeted with TALENs.
Another teleost tooth/fin enhancer has been described with overall similar expression patterns observed in this Bmp6 enhancer. In zebrafish, an FGF-responsive enhancer mediates Dlx2 expression in teeth and median and pectoral fins (Jackman and Stock, 2006). Additionally, in mice, a Bmp4 enhancer drives tooth epithelium and limb bud expression by responding to Pitx and Msx homeodomains (Jumlongras et al., 2012). The shared fin/limb and tooth expression domains of these cis-regulatory elements and the one described here suggest that fin and tooth development share multiple cis-regulatory networks, with at least three signaling pathways (FGF, Pitx/Msx, and TGFβ) involved in generating similar gene expression readouts in teeth and fins/limbs. Gene expression patterns of paired fins are thought to be co-opted from median fin expression domains in agnathans (Freitas et al., 2006). The Bmp6 enhancer presented here appears to be teleost-specific, as we did not find evidence of this conserved enhancer sequence in the genomes of lamprey, elephant shark, or spotted gar. Thus, our results suggest that teleosts may have secondarily coopted components of a gene regulatory network in developing median and pectoral fins and teeth.
Elucidating the cis-regulatory architecture of stickleback Bmp6 and evolved changes in Bmp6’s cis-regulatory architecture will help test the hypothesis that evolved changes in Bmp6 cis-regulation underlie the evolved increases in freshwater stickleback tooth number we previously described (Cleves et al., 2014). Although the 190 bp core Bmp6 enhancer presented here contains no nucleotide differences between low-toothed marine and high-toothed freshwater sticklebacks, several nucleotide differences exist in the sequence flanking the enhancer, which might contribute to the cis-regulatory differences observed between marine and freshwater alleles of Bmp6. Future studies will focus on whether these differences result in differential cis-regulatory activity between the marine and freshwater alleles of Bmp6.
Conservation and turnover of cis- and trans-regulatory information
It has been proposed that the cis-regulatory architecture of developmental control genes often consist of multiple independent modules, each of which drives expression in a particular tissue or cell type (Carroll, 2008; Stern, 2000). Because the Bmp6 enhancer drives multiple anatomical expression domains and is only partially conserved to zebrafish, we hypothesized that domains may have been sequentially added to the enhancer during teleost evolution, and that the different anatomical domains would be separable. Contrary to these predictions, our site directed mutagenesis and subcloning experiments of the stickleback Bmp6 enhancer appeared to affect all or none of the different expression domains, suggesting the different anatomical domains might not be separable and instead reflect ability to respond to a signal or signals present in multiple tissues.
Furthermore, enhancers from all four teleost species tested were sufficient to drive fin and tooth expression in zebrafish. However, the zebrafish enhancer, the most evolutionary divergent enhancer tested in this study, did not function robustly in sticklebacks, suggesting that the trans factors driving expression might have changed during the divergence of the two species. Similarly, testing a zebrafish Dlx2 tooth and fin enhancer in both zebrafish and Mexican tetra revealed that loss of oral Dlx2 expression in zebrafish is caused by changes in trans factors, as the Dlx2 zebrafish tooth enhancer is active in tetra oral teeth (Jackman and Stock, 2006). In both C. elegans and Drosophila, transgenic testing of cis-regulatory elements from two fly or worm species in both fly or worm species revealed that the greater the evolutionary distance separating two regulatory elements, the more likely upstream trans differences are to have evolved (Gordon and Ruvinsky, 2012). But, subtle changes in trans-acting factors can maintain similar expression patterns despite cis changes in divergent lineages (Barrière et al., 2012). Our results suggest a combination of conservation and divergence of trans factors, as stickleback sequence worked robustly in zebrafish, but zebrafish sequence was not functional in stickleback. Additionally, SB431542 treatment affected the stickleback enhancer in zebrafish more severely than in stickleback. Even at a low dose of SB431542 (25 µM), the enhancer was completely shut off in both epithelia and mesenchyme of zebrafish teeth (see Fig. 4E–F). This result supports potential trans regulatory divergence between stickleback and zebrafish, because it suggests that the enhancer’s expression may be more sensitive to TGFβ signaling in zebrafish than in stickleback.
A role for TGFβ in the regulation of BMPs
To our knowledge, this study is the first to support a role for TGFβ signaling in controlling Bmp signaling via a cis-regulatory input. Conditional deletion of Tgfbr1 (Alk5) in mouse neural crest lineages results in reduced expression of Bmp4 and delayed tooth initiation (Zhao et al., 2008); however, the mechanism of this interaction has not been described. Other studies have shown both positive and negative correlations between Bmp6 expression and TGFβ levels: Smad3 -/- chondrocytes have reduced Bmp6 expression (Li et al., 2006), whereas Bmp6 expression is increased in Smad3 -/- tendons undergoing tissue repair (Katzel et al., 2011). Our data suggest that in sticklebacks, TGFβ signaling activates Bmp6 expression in multiple tissues via a predicted Smad3 binding site. In teeth, blocking TGFβ signaling using the inhibitor SB431542 caused loss of epithelial reporter expression, but the effect on the mesenchymal expression was less severe (Fig. 4C, Fig. 5). The same pattern was observed in endogenous Bmp6 expression (Fig. 6O). This result suggests that epithelial and mesenchymal Bmp6 expression domains respond to partially different signaling pathways, with epithelial expression much more sensitive to TGFβ disruption.
We observed that a higher dose of TGFβ inhibitor SB431542 was required to shut off endogenous Bmp6 expression relative to expression driven solely by the 190 bp enhancer. While a 50 µM treatment almost completely eliminated enhancer expression (Fig. 4), at this dose we did not observe a strong difference in GFP expression driven by the reporter BAC. Only at the higher dose of 100 µM did we observe a change in BAC reporter expression and endogenous Bmp6 expression (Fig. 7). This finding suggests that in its native genomic context, the enhancer may be less sensitive to TGFβ signaling perturbations than when it is isolated in a reporter construct. There may be additional non-TGFβ regulatory elements that drive Bmp6 expression in the same tooth and fin domains such that a decrease in TGFβ signaling has a less obvious effect at lower doses. Furthermore, the effect of SB431542 treatment on endogenous Bmp6 expression and BAC reporter expression was not as dramatic as deletion of the Smad3 binding site with TALENs (compare Fig. 6 to Fig. 7). This finding suggests that other non-TGFβ factors may bind sequences immediately surrounding the Smad3 binding site to drive enhancer expression. However, the predicted Smad3 site is absolutely required, as loss of this site completely eliminates enhancer activity (Fig. 4J).
Combined effects of Wnt and TGFβ on tooth development
Although our site-directed mutagenesis experiment indicated that TCF/Lef predicted binding sites might be important for enhancer function (Fig. 3), pharmacological testing with XAV939 did not support the hypothesis that the enhancer requires Wnt signaling inputs for enhancer function. A stable line of zebrafish containing the TCF/Lef mutated reporter also drove robust reporter expression in fins and teeth, providing a second piece of evidence that the enhancer does not require Wnt input. This result was somewhat surprising, as the expression domains driven by the Bmp6 enhancer are similar to a TCF reporter zebrafish line (Shimizu et al., 2012). The reduction in activity seen from mutating the TCF/Lef sites may have been caused by other unknown binding sites overlapping the mutated base pairs, by inadvertently creating repressive motifs, or by somehow altering the binding of the Smad3 complex. The mutations may have affected the level, but not pattern, of GFP expression, making the construct appear less robust in our transient transgenic assay. We did note that combined treatment with XAV939 and SB431542 caused a slight decrease in mesenchymal tooth GFP expression (see insets of Fig. 5), however, this effect was less reproducible than the complete loss of epithelial expression seen upon SB431542 treatment alone.
The combination treatment with SB431542 and XAV939 did reduce tooth number in sticklebacks, suggesting that Wnt and TGFβ signaling pathways together are required for maintaining normal tooth development and patterning. In mice, as well as in diphyodont humans and polyphyodonts including snakes and alligators, Wnt signaling is required for tooth formation and replacement (Adaimy et al., 2007; Bohring et al., 2009; Gaete and Tucker, 2013; Genderen et al., 1994; Liu et al., 2008; Wu et al., 2013). In mice, TGFβ signaling is also required for tooth development (Ferguson et al., 1998, 2001; Oka et al., 2007). Antisense abrogation of both TGFB2 and TGFBRII in cultured mandibles resulted in accelerated tooth formation (Chai et al., 1994, 1999), however the TGFB2 knockout mouse has no reported tooth phenotype (Sanford et al., 1997). While the TGFBRII knockout dies prior to tooth formation (Oshima et al., 1996), conditional ablation in neural crest cells prevents terminal differentiation of odontoblasts (Oka et al., 2007), while conditional ablation in Osx-expressing odontoblasts revealed a necessary role for TGFBRII in molar root development (Wang et al., 2013). Furthermore, Wnt and TGFβ signaling are required to activate Eda and Edar in appropriate patterns in the developing tooth germs (Laurikkala et al., 2001). However, to our knowledge, this study is the first to show a partially redundant requirement for TGFβ and Wnt during tooth development, as only XAV939 and SB431542 doubly treated fish had reduced tooth numbers. Future studies of this enhancer will further test the hypothesis that this enhancer responds to TGFβ signaling to control Bmp6 expression during tooth and fin development.
Conclusions
We have identified a 190 base pair conserved enhancer required for tooth, fin, and other expression domains of stickleback Bmp6. Site directed mutagenesis and pharmacology experiments support the hypothesis that this enhancer responds to TGFβ signaling via a Smad3 binding site. Expression driven by this enhancer in tooth epithelial cells appears more sensitive to TGFβ levels than expression in tooth mesenchymal cells. To our knowledge, this is the first demonstration of a likely cis-regulatory link between TGFβ signaling and Bmp expression in teeth. In vivo deletion of this enhancer using TALENs caused severe disruption of Bmp6 expression in fins and tooth epithelia, suggesting this enhancer is required for normal expression patterns in a subset of Bmp6’s endogenous domains. Finally, we demonstrate that a combination of TGFβ signaling and Wnt signaling is required for normal tooth development in sticklebacks.
Supplementary Material
Acknowledgements
This work was supported by NIH R01 #DE021475. We thank David Kingsley for support and advice on BAC isolation, Tim Howes and David Kingsley for the generous gift of the Tol2/hsp70 backbone, Daniel Schlenk and Anita Kuepper for providing medaka specimens, Amy Strom and Katie Sieverman for assistance in cloning the TALENs, Natasha Naidoo for assistance in cloning the medaka reporter construct, and Lisa Kronstad for providing the site-directed mutagenesis protocol.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aberg T, Wozney J, Thesleff I. Expression patterns of bone morphogenetic proteins (Bmps) in the developing mouse tooth suggest roles in morphogenesis and cell differentiation. Dev. Dyn. 1997;210:383–396. doi: 10.1002/(SICI)1097-0177(199712)210:4<383::AID-AJA3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin’s finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Adaimy L, Chouery E, Mégarbané H, Mroueh S, Delague V, Nicolas E, Belguith H, de Mazancourt P, Mégarbané A. Mutation in WNT10A Is Associated with an Autosomal Recessive Ectodermal Dysplasia: The Odonto-onycho-dermal Dysplasia. Am. J. Hum. Genet. 2007;81:821–828. doi: 10.1086/520064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D, Karolak M, Robertson E, Oxburgh L. Control of kidney, eye and limb expression of Bmp7 by an enhancer element highly conserved between species. Dev. Biol. 2007;311:679–690. doi: 10.1016/j.ydbio.2007.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson RC, Streelman JT, Kocher TD, Yelick PC. Integration and evolution of the cichlid mandible: The molecular basis of alternate feeding strategies. Proc. Natl. Acad. Sci. U. S. A. 2005;102:16287–16292. doi: 10.1073/pnas.0506649102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andl T, Ahn K, Kairo A, Chu EY, Wine-Lee L, Reddy ST, Croft NJ, Cebra-Thomas JA, Metzger D, Chambon P, et al. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 2004;131:2257–2268. doi: 10.1242/dev.01125. [DOI] [PubMed] [Google Scholar]
- Andriopoulos B, Corradini E, Xia Y, Faasse SA, Chen S, Grgurevic L, Knutson MD, Pietrangelo A, Vukicevic S, Lin HY, et al. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat. Genet. 2009;41:482–487. doi: 10.1038/ng.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrière A, Gordon KL, Ruvinsky I. Coevolution within and between Regulatory Loci Can Preserve Promoter Function Despite Evolutionary Rate Acceleration. PLoS Genet. 2012;8:e1002961. doi: 10.1371/journal.pgen.1002961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bei M, Maas R. FGFs and BMP4 induce both Msx1-independent and Msx1-dependent signaling pathways in early tooth. Development. 1998;125:4325–4333. doi: 10.1242/dev.125.21.4325. [DOI] [PubMed] [Google Scholar]
- Bellusci S, Henderson R, Winnier G, Oikawa T, Hogan BL. Evidence from normal expression and targeted misexpression that bone morphogenetic protein (Bmp-4) plays a role in mouse embryonic lung morphogenesis. Development. 1996;122:1693–1702. doi: 10.1242/dev.122.6.1693. [DOI] [PubMed] [Google Scholar]
- Biggs LC, Mikkola ML. Early inductive events in ectodermal appendage morphogenesis. Semin. Cell Dev. Biol. 2014;25–26:11–21. doi: 10.1016/j.semcdb.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Bohring A, Stamm T, Spaich C, Haase C, Spree K, Hehr U, Hoffmann M, Ledig S, Sel S, Wieacker P, et al. WNT10A Mutations Are a Frequent Cause of a Broad Spectrum of Ectodermal Dysplasias with Sex-Biased Manifestation Pattern in Heterozygotes. Am. J. Hum. Genet. 2009;85:97–105. doi: 10.1016/j.ajhg.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev VA, Botchkareva NV, Roth W, Nakamura M, Chen LH, Herzog W, Lindner G, McMahon JA, Peters C, Lauster R, et al. Noggin is a mesenchymally derived stimulator of hair-follicle induction. Nat. Cell Biol. 1999;1:158–164. doi: 10.1038/11078. [DOI] [PubMed] [Google Scholar]
- Calle-Mustienes E de la, Feijóo CG, Manzanares M, Tena JJ, Rodríguez-Seguel E, Letizia A, Allende ML, Gómez-Skarmeta JL. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 2005;15:1061–1072. doi: 10.1101/gr.4004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evo-Devo and an Expanding Evolutionary Synthesis: A Genetic Theory of Morphological Evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. gkr. 2011:218. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Mah A, Crohin C, Groff S, Bringas P, Le T, Santos V, Slavkin HC. Specific transforming growth factor-beta subtypes regulate embryonic mouse Meckel’s cartilage and tooth development. Dev. Biol. 1994;162:85–103. doi: 10.1006/dbio.1994.1069. [DOI] [PubMed] [Google Scholar]
- Chai Y, Zhao J, Mogharei A, Xu B, Bringas P, Jr, Shuler C, Warburton D. Inhibition of transforming growth factor-β type II receptor signaling accelerates tooth formation in mouse first branchial arch explants. Mech. Dev. 1999;86:63–74. doi: 10.1016/s0925-4773(99)00112-4. [DOI] [PubMed] [Google Scholar]
- Chandler RL, Chandler KJ, McFarland KA, Mortlock DP. Bmp2 Transcription in Osteoblast Progenitors Is Regulated by a Distant 3′ Enhancer Located 156.3 Kilobases from the Promoter. Mol. Cell. Biol. 2007;27:2934–2951. doi: 10.1128/MCB.01609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Bei M, Woo I, Satokata I, Maas R. Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development. 1996;122:3035–3044. doi: 10.1242/dev.122.10.3035. [DOI] [PubMed] [Google Scholar]
- Cleves PA, Ellis NA, Jimenez MT, Nunez SM, Schluter D, Kingsley DM, Miller CT. Evolved tooth gain in sticklebacks is associated with a cis-regulatory allele of Bmp6. Proc. Natl. Acad. Sci. 2014;111:13912–13917. doi: 10.1073/pnas.1407567111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassule HR, McMahon AP. Analysis of Epithelial-Mesenchymal Interactions in the Initial Morphogenesis of the Mammalian Tooth. Dev. Biol. 1998;202:215–227. doi: 10.1006/dbio.1998.8992. [DOI] [PubMed] [Google Scholar]
- Dathe K, Kjaer KW, Brehm A, Meinecke P, Nürnberg P, Neto JC, Brunoni D, Tommerup N, Ott CE, Klopocki E, et al. Duplications Involving a Conserved Regulatory Element Downstream of BMP2 Are Associated with Brachydactyly Type A2. Am. J. Hum. Genet. 2009;84:483–492. doi: 10.1016/j.ajhg.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendooven A, van Oostrom O, van der Giezen DM, Willem Leeuwis J, Snijckers C, Joles JA, Robertson EJ, Verhaar MC, Nguyen TQ, Goldschmeding R. Loss of Endogenous Bone Morphogenetic Protein-6 Aggravates Renal Fibrosis. Am. J. Pathol. 2011;178:1069–1079. doi: 10.1016/j.ajpath.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, VanDyk JK, Bogdanove AJ. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–W122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley AT, Godin RE, Robertson EJ. Interaction between FGF and BMP signaling pathways regulates development of metanephric mesenchyme. Genes Dev. 1999;13:1601–1613. doi: 10.1101/gad.13.12.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, Messeguer X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng JQ, Zhang J, Tan X, Lu Y, Guo D, Harris SE. Identification of Cis-DNA Regions Controlling Bmp4 Expression during Tooth Morphogenesis in vivo. J. Dent. Res. 2002;81:6–10. doi: 10.1177/002203450208100103. [DOI] [PubMed] [Google Scholar]
- Ferguson CA, Tucker AS, Christensen L, Lau AL, Matzuk MM, Sharpe PT. Activin is an essential early mesenchymal signal in tooth development that is required for patterning of the murine dentition. Genes Dev. 1998;12:2636–2649. doi: 10.1101/gad.12.16.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson CA, Tucker AS, Heikinheimo K, Nomura M, Oh P, Li E, Sharpe PT. The role of effectors of the activin signalling pathway, activin receptors IIA and IIB, and Smad2, in patterning of tooth. Development. 2001;128:4605–4613. doi: 10.1242/dev.128.22.4605. [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat. Protoc. 2006;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- Fraser GJ, Bloomquist RF, Streelman JT. Common developmental pathways link tooth shape to regeneration. Dev. Biol. 2013;377:399–414. doi: 10.1016/j.ydbio.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas R, Zhang G, Cohn MJ. Evidence that mechanisms of fin development evolved in the midline of early vertebrates. Nature. 2006;442:1033–1037. doi: 10.1038/nature04984. [DOI] [PubMed] [Google Scholar]
- Fujimori S, Novak H, Weissenböck M, Jussila M, Gonçalves A, Zeller R, Galloway J, Thesleff I, Hartmann C. Wnt/β-catenin signaling in the dental mesenchyme regulates incisor development by regulating Bmp4. Dev. Biol. 2010;348:97–106. doi: 10.1016/j.ydbio.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaete M, Tucker AS. Organized Emergence of Multiple-Generations of Teeth in Snakes Is Dysregulated by Activation of Wnt/Beta-Catenin Signalling. PLoS ONE. 2013;8:e74484. doi: 10.1371/journal.pone.0074484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genderen C, van Okamura RM, Fariñas I, Quo RG, Parslow TG, Bruhn L, Grosschedl R. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- Gordon KL, Ruvinsky I. Tempo and Mode in Evolution of Transcriptional Regulation. PLoS Genet. 2012;8:e1002432. doi: 10.1371/journal.pgen.1002432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, Sulem P, Thorlacius S, Gylfason A, Steinberg S, et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- Guenther C, Pantalena-Filho L, Kingsley DM. Shaping Skeletal Growth by Modular Regulatory Elements in the Bmp5 Gene. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan BL. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- Houlston RS, Webb E, Broderick P, Pittman AM, Di Bernardo MC, Lubbe S, Chandler I, Vijayakrishnan J, Sullivan K, Penegar S, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat. Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S-MA, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- Inman GJ, Nicolás FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. SB-431542 Is a Potent and Specific Inhibitor of Transforming Growth Factor-β Superfamily Type I Activin Receptor-Like Kinase (ALK) Receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- Jackman WR, Stock DW. Transgenic analysis of Dlx regulation in fish tooth development reveals evolutionary retention of enhancer function despite organ loss. Proc. Natl. Acad. Sci. 2006;103:19390–19395. doi: 10.1073/pnas.0609575103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman WR, Davies SH, Lyons DB, Stauder CK, Denton-Schneider BR, Jowdry A, Aigler SR, Vogel SA, Stock DW. Manipulation of Fgf and Bmp signaling in teleost fishes suggests potential pathways for the evolutionary origin of multicuspid teeth. Evol. Dev. 2013;15:107–118. doi: 10.1111/ede.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumlongras D, Lachke SA, O’Connell DJ, Aboukhalil A, Li X, Choe SE, Ho JWK, Turbe-Doan A, Robertson EA, Olsen BR, et al. An Evolutionarily Conserved Enhancer Regulates Bmp4 Expression in Developing Incisor and Limb Bud. PLoS ONE. 2012;7:e38568. doi: 10.1371/journal.pone.0038568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H-S, Francis-West PH, Widelitz RB, Jiang T-X, Ting-Berreth S, Tickle C, Wolpert L, Chuong C-M. Local Inhibitory Action of BMPs and Their Relationships with Activators in Feather Formation: Implications for Periodic Patterning. Dev. Biol. 1998;196:11–23. doi: 10.1006/dbio.1998.8850. [DOI] [PubMed] [Google Scholar]
- Justice CM, Yagnik G, Kim Y, Peter I, Jabs EW, Erazo M, Ye X, Ainehsazan E, Shi L, Cunningham ML, et al. A genome-wide association study identifies susceptibility loci for nonsyndromic sagittal craniosynostosis near BMP2 and within BBS9. Nat. Genet. 2012;44:1360–1364. doi: 10.1038/ng.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzel EB, Wolenski M, Loiselle AE, Basile P, Flick LM, Langstein HN, Hilton MJ, Awad HA, Hammert WC, O’Keefe RJ. Impact of Smad3 loss of function on scarring and adhesion formation during tendon healing. J. Orthop. Res. Off. Publ. Orthop. Res. Soc. 2011;29:684–693. doi: 10.1002/jor.21235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh KD, Evans AR, Jernvall J. Predicting evolutionary patterns of mammalian teeth from development. Nature. 2007;449:427–432. doi: 10.1038/nature06153. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A Transposon-Mediated Gene Trap Approach Identifies Developmentally Regulated Genes in Zebrafish. Dev. Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Kingsley DM. What do BMPs do in mammals? Clues from the mouse short-ear mutation. Trends Genet. 1994;10:16–21. doi: 10.1016/0168-9525(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Laurikkala J, Mikkola M, Mustonen T, Åberg T, Koppinen P, Pispa J, Nieminen P, Galceran J, Grosschedl R, Thesleff I. TNF Signaling via the Ligand-Receptor Pair Ectodysplasin and Edar Controls the Function of Epithelial Signaling Centers and Is Regulated by Wnt and Activin during Tooth Organogenesis. Dev. Biol. 2001;229:443–455. doi: 10.1006/dbio.2000.9955. [DOI] [PubMed] [Google Scholar]
- Li T-F, Darowish M, Zuscik MJ, Chen D, Schwarz EM, Rosier RN, Drissi H, O’Keefe RJ. Smad3-deficient chondrocytes have enhanced BMP signaling and accelerated differentiation. J. Bone Miner. Res. 2006;21:4–16. doi: 10.1359/JBMR.050911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Chu EY, Watt B, Zhang Y, Gallant NM, Andl T, Yang SH, Lu M-M, Piccolo S, Schmidt-Ullrich R, et al. Wnt/β-catenin signaling directs multiple stages of tooth morphogenesis. Dev. Biol. 2008;313:210–224. doi: 10.1016/j.ydbio.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Sun X, Braut A, Mishina Y, Behringer RR, Mina M, Martin JF. Distinct functions for Bmp signaling in lip and palate fusion in mice. Development. 2005;132:1453–1461. doi: 10.1242/dev.01676. [DOI] [PubMed] [Google Scholar]
- Lubbe SJ, Pittman AM, Olver B, Lloyd A, Vijayakrishnan J, Naranjo S, Dobbins S, Broderick P, Gómez-Skarmeta JL, Houlston RS. The 14q22.2 colorectal cancer variant rs4444235 shows cis-acting regulation of BMP4. Oncogene. 2012;31:3777–3784. doi: 10.1038/onc.2011.564. [DOI] [PubMed] [Google Scholar]
- Marinić M, Aktas T, Ruf S, Spitz F. An Integrated Holo-Enhancer Unit Defines Tissue and Gene Specificity of the Fgf8 Regulatory Landscape. Dev. Cell. 2013;24:530–542. doi: 10.1016/j.devcel.2013.01.025. [DOI] [PubMed] [Google Scholar]
- Massagué J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messeguer X, Escudero R, Farré D, Núñez O, Martınez J, Albà MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- Mou C, Jackson B, Schneider P, Overbeek PA, Headon DJ. Generation of the primary hair follicle pattern. Proc. Natl. Acad. Sci. 2006;103:9075–9080. doi: 10.1073/pnas.0600825103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou C, Pitel F, Gourichon D, Vignoles F, Tzika A, Tato P, Yu L, Burt DW, Bed’hom B, Tixier-Boichard M, et al. Cryptic Patterning of Avian Skin Confers a Developmental Facility for Loss of Neck Feathering. PLoS Biol. 2011;9:e1001028. doi: 10.1371/journal.pbio.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, Wainwright PC, Friedman M, Smith WL. Resolution of ray-finned fish phylogeny and timing of diversification. Proc. Natl. Acad. Sci. 2012;109:13698–13703. doi: 10.1073/pnas.1206625109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubüser A, Peters H, Balling R, Martin GR. Antagonistic Interactions between FGF and BMP Signaling Pathways: A Mechanism for Positioning the Sites of Tooth Formation. Cell. 1997;90:247–255. doi: 10.1016/s0092-8674(00)80333-5. [DOI] [PubMed] [Google Scholar]
- Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37:D77–D82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X, Luukko K, Kettunen P. BMP signalling in craniofacial development. Int. J. Dev. Biol. 2006:50. doi: 10.1387/ijdb.052101xn. [DOI] [PubMed] [Google Scholar]
- O’Connell DJ, Ho JWK, Mammoto T, Turbe-Doan A, O’Connell JT, Haseley PS, Koo S, Kamiya N, Ingber DE, Park PJ, et al. A Wnt-bmp feedback circuit controls intertissue signaling dynamics in tooth organogenesis. Sci. Signal. 2012;5:ra4. doi: 10.1126/scisignal.2002414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka S, Oka K, Xu X, Sasaki T, Bringas P, Jr, Chai Y. Cell autonomous requirement for TGF-β signaling during odontoblast differentiation and dentin matrix formation. Mech. Dev. 2007;124:409–415. doi: 10.1016/j.mod.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima M, Oshima H, Taketo MM. TGF-beta receptor type II deficiency results in defects of yolk sac hematopoiesis and vasculogenesis. Dev. Biol. 1996;179:297–302. doi: 10.1006/dbio.1996.0259. [DOI] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol. 2010;20:1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pregizer S, Mortlock DP. Control of BMP gene expression by long-range regulatory elements. Cytokine Growth Factor Rev. 2009;20:509–515. doi: 10.1016/j.cytogfr.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MT, LaBrie S, McPherson J, Stanton VP. Screening large-insert libraries by hybridization. In: Dracopoli NC, Haines JL, Korf B, editors. Current Protocols in Human Genetics. New York: John Wiley and Sons; 1999. pp. 5.6.1–5.6.52. [Google Scholar]
- Sanford LP, Ormsby I, Groot ACG, Sariola H, Friedman R, Boivin GP, Cardell EL, Doetschman T. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development. 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer N, Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech. Dev. 1999;80:153–158. doi: 10.1016/s0925-4773(98)00209-3. [DOI] [PubMed] [Google Scholar]
- Shi M, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, Wu T, Murray T, Redett RJ, Wilcox AJ, et al. Genome wide study of maternal and parent-of-origin effects on the etiology of orofacial clefts. Am. J. Med. Genet. A. 2012;158A:784–794. doi: 10.1002/ajmg.a.35257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N, Kawakami K, Ishitani T. Visualization and exploration of Tcf/Lef function using a highly responsive Wnt/β-catenin signaling-reporter transgenic zebrafish. Dev. Biol. 2012;370:71–85. doi: 10.1016/j.ydbio.2012.07.016. [DOI] [PubMed] [Google Scholar]
- Solloway MJ, Dudley AT, Bikoff EK, Lyons KM, Hogan BL, Robertson EJ. Mice lacking Bmp6 function. Dev. Genet. 1998;22:321–339. doi: 10.1002/(SICI)1520-6408(1998)22:4<321::AID-DVG3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Stern DL. Perspective: Evolutionary Developmental Biology and the Problem of Variation. Evolution. 2000;54:1079–1091. doi: 10.1111/j.0014-3820.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- Sun Z, Jin P, Tian T, Gu Y, Chen Y-G, Meng A. Activation and roles of ALK4/ALK7-mediated maternal TGFβ signals in zebrafish embryo. Biochem. Biophys. Res. Commun. 2006;345:694–703. doi: 10.1016/j.bbrc.2006.04.148. [DOI] [PubMed] [Google Scholar]
- Suster ML, Abe G, Schouw A, Kawakami K. Transposon-mediated BAC transgenesis in zebrafish. Nat. Protoc. 2011;6:1998–2021. doi: 10.1038/nprot.2011.416. [DOI] [PubMed] [Google Scholar]
- Swarup H. Stages in the Development of the Stickleback Gasterosteus aculeatus (L.) J. Embryol. Exp. Morphol. 1958;6:373–383. [PubMed] [Google Scholar]
- Urasaki A, Morvan G, Kawakami K. Functional Dissection of the Tol2 Transposable Element Identified the Minimal cis-Sequence and a Highly Repetitive Sequence in the Subterminal Region Essential for Transposition. Genetics. 2006;174:639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainio S, Karavanova I, Jowett A, Thesleff I. Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell. 1993;75:45–58. [PubMed] [Google Scholar]
- Villefranc JA, Amigo J, Lawson ND. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev. Dyn. 2007;236:3077–3087. doi: 10.1002/dvdy.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cox MK, Coricor G, MacDougall M, Serra R. Inactivation of Tgfbr2 in Osterix-Cre expressing dental mesenchyme disrupts molar root formation. Dev. Biol. 2013;382:27–37. doi: 10.1016/j.ydbio.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A guide for the Laboratory Use of Zebrafish (Danio rerio) 5th Edition. Eugene, OR: University of Oregon Press; 2007. [Google Scholar]
- Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Chu AY, Estrada K, Luan J, Kutalik Z, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Wu X, Jiang T-X, Elsey RM, Temple BL, Divers SJ, Glenn TC, Yuan K, Chen M-H, Widelitz RB, et al. Specialized stem cell niche enables repetitive renewal of alligator teeth. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E2009–E2018. doi: 10.1073/pnas.1213202110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Oka K, Bringas P, Kaartinen V, Chai Y. TGF-β type I receptor Alk5 regulates tooth initiation and mandible patterning in a type II receptor-independent manner. Dev. Biol. 2008;320:19–29. doi: 10.1016/j.ydbio.2008.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Zhang Z, Song Y, Zhang X, Zhang Y, Hu Y, Fromm SH, Chen Y. Transgenically ectopic expression of Bmp4 to the Msx1 mutant dental mesenchyme restores downstream gene expression but represses Shh and Bmp2 in the enamel knot of wild type tooth germ. Mech. Dev. 2000;99:29–38. doi: 10.1016/s0925-4773(00)00467-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.