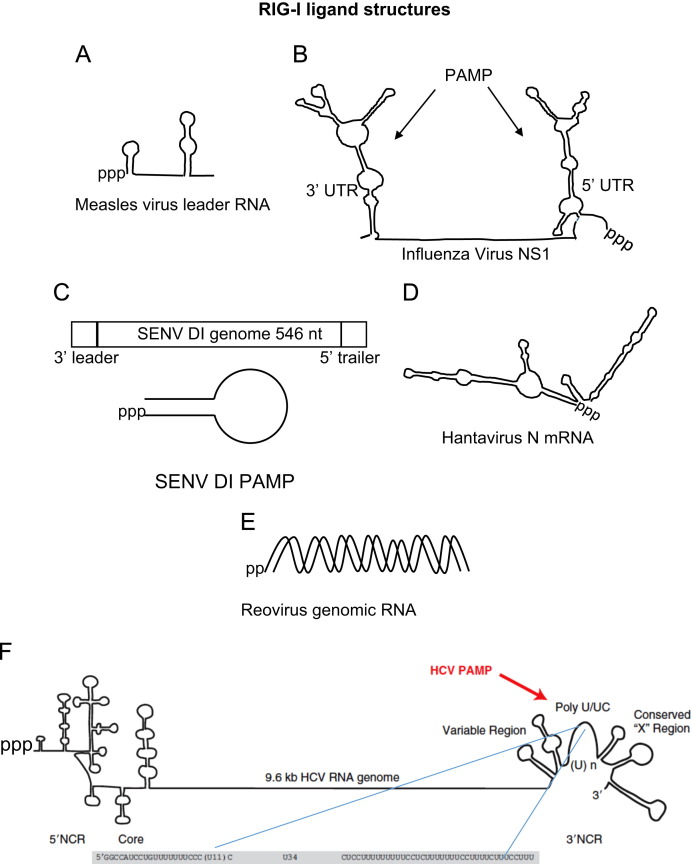
Fig. 2.
Examples of known RIG-I PAMP RNAs with models of specific structures shown. (A) RNAfold predicted structure of measles virus leader RNA using Edmonson B strain sequence. (B) Influenza virus PAMPs found at genomic 5′ and 3′ nontranslated regions of the NS1 segment (adapted from Davis et al. (2012)). (C) Sendai virus defective-interfering (DI) particle structure (adapted from Martinez-Gil et al. (2013)). (D) RNAfold predicted structure of Hantavirus nucleoprotein mRNA using TPM strain sequence. (E) Reovirus genomic dsRNA with 5′pp motif identified as RIG-I PAMP in Goubau et al. (2014). (F) Hepatitis C virus genome with 3′ nontranslated region and poly-U/UC PAMP highlighted (adapted from Horner and Gale (2013)).