Abstract
Influenza A viruses cause respiratory infections that range from asymptomatic to deadly in humans. Widespread outbreaks (pandemics) are attributable to ‘novel’ viruses that possess a viral hemagglutinin (HA) gene to which humans lack immunity. After a pandemic, these novel viruses form stable virus lineages in humans and circulate until they are replaced by other novel viruses. The factors and mechanisms that facilitate virus transmission among hosts and the establishment of novel lineages are not completely understood, but the HA and basic polymerase 2 (PB2) proteins are thought to play essential roles in these processes by enabling avian influenza viruses to infect mammals and replicate efficiently in their new host. Here, we summarize our current knowledge of the contributions of HA, PB2, and other viral components to virus transmission and the formation of new virus lineages.
Keywords: Influenza virus, transmission, virus lineage, HA, PB2, NA, receptor-binding, gain-of-function
On average, influenza viruses infect 5%–10% of human populations each year (http://www.who.int/mediacentre/factsheets/fs211/en/); these percentages can be considerably higher in certain geographical areas or age groups, as well as when novel influenza viruses encounter a human population that lacks adequate neutralizing immune responses against such novel viruses. The loss of life and economic costs are considerable during annual influenza virus epidemics, and can be tremendous during worldwide outbreaks of novel strains (pandemics). Pandemics may also cause heightened public apprehension, resulting in the disruption of daily life. In addition to the impact on human health, outbreaks of influenza viruses can also cause considerable losses to the poultry, swine, and race horse industries.
The Orthomyxoviridae family of viruses comprises five genera, but only two are of medical relevance in humans, namely, influenza A and B viruses. Influenza A viruses are further divided into subtypes, based on the antigenic properties of the surface glycoproteins hemagglutinin (HA) and neuraminidase (NA), the two major viral antigens. Wild waterfowl are believed to be the reservoir of influenza A viruses of the H1–H16 HA and the N1–N9 NA subtypes. To date, genetic material of influenza viruses of the H17N10 and H18N11 subtypes has been detected only in bats (1, 2). From their natural reservoir, influenza A viruses can be transmitted to numerous other species, primarily poultry, pigs, and humans. In humans, only viruses of the H1N1, H2N2, and H3N2 subtypes have circulated for extended periods. Avian influenza viruses of the H5N1 and H7N9 subtypes have infected hundreds of people since their emergence in 1997 and 2013, respectively, but have failed to spread efficiently among humans. In addition, there have been reports of isolated cases of human infections with viruses of several other subtypes, namely H9N2, H6N1, H7N7, H10N8, H7N2, and H7N3 (3-10).
The influenza virus life cycle is initiated by the binding of viral HA to receptors on host cells. After endocytosis and HA-mediated fusion of the viral and cellular membranes, viral ribonucleoprotein complexes are released into the cytoplasm, transported to the nucleus, and replicated and transcribed by the viral polymerase complex. Newly formed viral ribonucleoprotein complexes and structural proteins are transported to the plasma membrane, where the new viruses are formed and bud (a detailed description of the viral life cycle can be found in reference (11)).
We do not yet have a complete understanding of the factors and mechanisms that are essential for influenza virus transmission, cause pandemics, and lead to the establishment of novel lineages. In the broadest terms, influenza virus evolution is driven by two mechanisms: mutations in the viral genome, and reassortment, which is the rearrangement of the eight influenza A viral RNA (vRNA) segments in cells infected with at least two different viruses. Here, we review how these mechanisms affect the transmissibility of influenza A viruses.
Establishment of Novel Influenza A Virus Lineages in Humans
In 1918, a novel influenza A virus that transmitted efficiently among humans caused a pandemic of unparalleled scale, killing an estimated 50–100 million people worldwide. Taubenberger and colleagues reconstituted the sequence of this pandemic virus from historic samples (reviewed in (12)). Based on the analysis of the viral sequences, the pandemic virus most likely originated from an avian host. Descendants of the pandemic virus formed a new virus lineage in humans and circulated until 1957, when they were replaced by a novel influenza virus of the H2N2 subtype, which caused the ‘Asian’ influenza. This virus was a reassortant with HA, NA, and polymerase PB1 vRNA segments derived from an avian influenza virus, and the remaining five vRNA segments originating from the previously circulating H1N1 viruses. This novel virus became established in human populations and circulated for the next decade. In 1968, another reassortant virus caused the ‘Hong Kong’ influenza. It possessed six vRNA segments from the previously circulating H2N2 viruses, whereas its HA and PB1 vRNA segments were derived from avian H3 influenza viruses. After causing a pandemic, the novel H3N2 viruses became established in humans and are still circulating in human populations to this day. In 1977, H1N1 viruses closely related to those circulating in humans in the 1950s reemerged; the origin of these reemerging viruses is unknown. The H1N1 viruses again formed a stable lineage in humans and co-circulated with human H3N2 viruses until they were replaced by a novel pandemic H1N1 virus in 2009. This most recent pandemic resulted from multiple reassortment events, which most likely occurred in pigs: in the late 1990s, novel ‘triple reassortant’ H1N2 viruses emerged in North American pigs. These viruses possessed PB2 and PA polymerase vRNA segments derived from avian influenza viruses; nucleoprotein (NP), matrix (M), and nonstructural (NS) vRNA segments derived from previously circulating H1N1 swine influenza viruses; and HA, NA, and PB1 vRNA segments derived from human influenza viruses (13-16). These ‘triple reassortant’ viruses caused sporadic infections (17-20) but did not become adapted to humans. In 2009, however, the NA and M vRNA segments of a Eurasian avian-like swine virus reassorted with the remaining six vRNA segments of the ‘triple reassortant’ swine viruses, resulting in the 2009 pandemic H1N1 virus (reviewed in (21)). This virus has now established a stable lineage in humans and currently co-circulates with human H3N2 influenza viruses.
Sporadic Infections of Humans with Swine or Avian Influenza Viruses
In 1976, a classic H1N1 swine virus was isolated from five soldiers at Fort Dix, New Jersey, USA (22-25). Serological studies demonstrated that more than 500 people had been infected with this virus (22, 24, 26), suggesting that it was capable of human-to-human transmission. Yet, this virus did not establish a stable lineage in humans. Since August 2011, more than 340 people in North America have been infected with so-called ‘H3N2v’ (H3N2 variant) swine viruses. These viruses share the M vRNA segment with 2009 pandemic H1N1 viruses, but possesses HA and NA vRNA segments derived from human H3N2 viruses that were introduced into pigs in the mid-1990s (27-30). Epidemiological data suggest pigs as the likely source of human H3N2v infections. Most reported cases have been in children, presumably because they were not infected with the human H3N2 viruses that circulated in the mid-1990's and therefore lacked protective immunity against them. No sustained human-to-human transmission of this virus has been reported to date.
Generally, infections of humans with avian influenza viruses are rare; however, in recent years, avian influenza A viruses of the H5N1 and H7N9 subtypes have infected hundreds of people with case fatality rates of ~60% and ~30%, respectively.
The transmission of highly pathogenic avian H5N1 viruses to humans was first reported in 1997 in Hong Kong (31-33). The culling of birds in live poultry markets in Hong Kong brought a temporary end to the human infections, but new cases were reported in 2003. Since then, highly pathogenic avian H5N1 influenza viruses have spread across three continents, have become enzootic in poultry populations in parts of Southeast Asia, the Middle East, and perhaps also Indonesia, and have infected 694 people as of January 6, 2015, 492 (71%) of whom died (http://www.who.int/influenza/human_animal_interface/EN_GIP_20150106CumulativeNumberH 5N1cases.pdf?ua=1). Isolated cases of suspected human-to-human transmission have been reported, but sustained transmission chains in humans—which could lead to a pandemic—have not occurred.
Infections of humans with influenza viruses of the H7 subtype are typically self-limiting and associated with conjunctivitis and/or mild respiratory disease (reviewed in (34)), although a fatal human infection with an H7N7 virus occurred in 2003 in the Netherlands (9, 10). Since the spring of 2013, however, more than 450 human infections with a novel influenza virus of the H7N9 subtype have been documented. Epidemiologic and genetic data suggest poultry as the likely source of these infections. The internal genes of the H7N9 virus closely resemble those of H9N2 poultry viruses that have been circulating in China in recent years; the H7 HA and N9 NA vRNA segments most likely originated from H7 viruses circulating in ducks in southern China in 2010 and 2011, and from duck or wild bird N9 viruses, respectively (35-43). Thus, the H7N9 viruses were most likely created by several reassortment events among avian viruses. To date, no sustained transmission of H7N9 viruses among humans has been reported.
The Role of HA Receptor-Binding Specificity in Influenza A Virus Transmission
Infections of humans with avian or swine influenza viruses occur sporadically, but typically do not result in the establishment of new lineages. Numerous studies over the last few decades have identified the viral HA protein as the major host restriction factor that limits interspecies transmission and the establishment of virus lineages in new hosts.
Influenza viruses bind to sialyloligosaccharides on glycoproteins or glycolipids on the surface of host cells to infect them. Sialic acids can be linked to the penultimate galactose through different linkages, such as an α2,3- or an α2,6-linkage (Siaα2,3Gal or Siaα2,6Gal). The epithelial cells in the duck intestine express primarily Siaα2,3Gal (44), and influenza A viruses circulating in wild waterfowl efficiently bind to this type of sialic acid (45, 46). By contrast, epithelial cells in the human trachea contain predominately Siaα2,6Gal, whereas Siaα2,3Gal is found on certain alveolar cells (47-53). Influenza viruses circulating in humans have therefore adapted to bind efficiently to Siaα2,6Gal (54). The epithelial cells of the pig trachea contain both Siaα2,3Gal and Siaα2,6Gal (55-57), which may explain why pigs can be infected efficiently by human and avian influenza viruses; consequently, pigs may serve as ‘mixing vessels’ for the reassortment of avian, swine, and human influenza viruses, an event that likely led to the pandemic of 2009. Both types of sialic acid are also found on epithelial cells in the gastrointestinal and respiratory tract of chickens and other terrestrial poultry (58), consistent with the finding that influenza viruses isolated from terrestrial poultry can have dual avian/human-type receptor-binding specificity.
For this reason, it has been proposed that terrestrial poultry may be an intermediate host, facilitating the adaptation of avian influenza viruses to mammals.
Receptor-binding specificity and transmissibility of H1 influenza viruses
For H1 HAs, the amino acids at two positions, 190 and 225, define the binding preference for Siaα2,3Gal or Siaα2,6Gal (54, 59); note that all HA amino acid positions listed in this article refer to the numbering scheme of H3 HA as described by Burke & Smith (60). Glutamic acid and glycine at these positions confer efficient binding to avian-type receptors, whereas aspartic acid at these positions allows efficient binding to Siaα2,6Gal. The A/South Carolina/1/1918 (H1N1) pandemic virus encodes the human-type residues at positions 190 and 225 of its HA protein (i.e., HA-190D, HA-225D), bound to Siaα2,6Gal (59), and transmitted efficiently via respiratory droplets among ferrets (61); resolution of its crystal structure provided further insights into its human-type receptor-binding preference (62). By contrast, the HA protein of A/New York/1/1918 pandemic virus (encoding the human-type HA-190D and avian-type HA-225G residues) bound to both Siaα2,6Gal and Siaα2,3Gal (59); this virus transmitted less efficiently via respiratory droplets in ferrets than did A/South Carolina/1/18 virus (61). A mutant 1918 HA with the avian-type HA-190E and HA-225G residues displayed avian-type receptor-binding specificity (59) and did not facilitate transmission among ferrets (61). Hence, van Hoeven et al. (63) asked whether 1918 pandemic viral genes could confer respiratory droplet transmissibility to an avian influenza virus that typically lacks this property. They found that the HA gene of the 1918 pandemic virus alone was not sufficient to confer virus transmissibility in ferrets; the PB2 gene of the 1918 pandemic virus was also needed.
Recently, Watanabe et al. (64) generated a ‘1918-like avian’ virus composed of avian influenza vRNA segments with high homology to the 1918 pandemic virus. This ‘1918-like avian’ virus was more pathogenic in mice and in ferrets than a natural avian influenza virus but did not transmit via respiratory droplets among ferrets (64). However, a mutant variant of this virus possessing seven mutations in its HA and polymerase proteins (namely, HA-E89D, -S113N, -E190D, -G225D; PB2-E627K, -A684D; PA-V253M) transmitted among ferrets via respiratory droplets (64). The HA-E190D/G225D and PB2-E627K changes were introduced to confer human-type receptor-binding specificity (see above) and efficient replication in mammals (see ‘The Role of Polymerase Proteins in Influenza A Virus Transmission’), whereas the other mutations emerged during virus amplification in cell culture or replication in ferrets. This study demonstrated that contemporary avian influenza viruses encoding viral proteins similar to those of the 1918 pandemic virus may acquire the ability to transmit among mammals; moreover, this study again demonstrated the important role of HA and PB2 in virus transmissibility.
The HA proteins of the 2009 pandemic H1N1 viruses encode human-type amino acids at positions 190 and 225 and bind primarily to Siaα2,6Gal (65-67), although one study reported that these viruses have dual avian/human-type receptor-binding specificity (68). X-ray crystallographic analysis of 2009 pandemic H1N1 virus HA proteins has provided a structural explanation for human-type receptor-binding specificity (66, 69). Consistent with their human-type receptor-binding specificity, 2009 pandemic H1N1 viruses transmitted efficiently among ferrets and guinea pigs in direct contact and respiratory droplet transmission models (21, 70-73). Some 2009 pandemic H1N1 viruses encode the avian virus-type glycine residue at position 225, which confers dual avian/human-type receptor-binding specificity and appears to correlate with increased severity of disease (74-82). However, this mutation has not increased in frequency, suggesting that viruses that carry it do not transmit efficiently among humans.
Receptor-binding specificity and transmissibility of H7 influenza viruses
The human-type receptor-binding specificity of H3 HAs is conferred by leucine and serine at positions 226 and 228, in contrast to glutamine and glycine at these positions, which facilitate efficient binding to avian-type receptors (54, 83, 84). H3 and H7 HAs are phylogenetically closely related, suggesting that the amino acids at position 226 and 228 may also determine the receptor-binding specificity of H7 influenza viruses. The H7N7 influenza virus isolated from the fatal case in the Netherlands in 2003 (A/Netherlands/219/2003, H7N7) encoded avian-type amino acids at positions 226 and 228, displayed avian-type receptor-binding specificity (85) and did not transmit among ferrets (85). A virus isolated during the same outbreak from a person who developed conjunctivitis (A/Netherlands/230/2003, H7N7) was transmitted to cage mates (86); the HA proteins of these two viruses differ by three amino acids, but their contribution to transmissibility is currently unknown. Some North American H7 viruses display increased binding to human-type receptors and/or transmit to co-housed ferrets, but not to animals in adjacent cages (85-88).
The H7N9 viruses that emerged in China in 2013 encode human virus-type leucine at position 226, but the avian-type amino acid at position 228. Most H7N9 viruses also encode valine at HA position 186, which is known to affect the receptor-binding specificity of H7 HAs (88-90). One H7N9 isolate (A/Shanghai/1/2013) encodes HA-138S, which increases the binding of pig H5N1 influenza viruses to Siaα2,6Gal (91). It is, therefore, not surprising that the H7N9 viruses bind to both Siaα2,3Gal and Siaα2,6Gal, although differences have been reported depending on the isolate and the nature of the sample (i.e., purified HA protein or whole virus) (92-98). The X-ray crystallographic structure of A/Anhui/1/2013 HA has been solved in complex with human- and avian-type receptors (94, 96): the human virus-type HA-226L residue creates a non-polar binding site for the human-type receptor, whereas HA-186V increases the hydrophobicity of the binding site. However, the orientation of the human-type receptor in the binding pocket differed from that observed for pandemic viruses and an H5 HA virus that conferred transmissibility in humans.
The dual avian/human virus receptor-binding specificity of H7N9 viruses likely explains their respiratory droplet transmissibility, although these viruses transmit less efficiently than human influenza viruses (95, 99-104). In guinea pigs, efficient contact transmission to cage mates (105) and respiratory droplet transmission (106) have been detected. By contrast, no H7N9 virus transmission occurred between pigs and ferrets in an experimental setting (104). Sequential passages of H7N9 viruses in ferrets does not increase the respiratory droplet transmissibility of H7N9 viruses (101). A single passage of an avian H7N9 virus (A/chicken/Zhejiang/DTID-ZJU01/2013) in pigs resulted in a virus with mutations in the HA, NA, M, and NS vRNA segments, which conferred enhanced binding to human-type receptors and high virus titers in the lungs of infected pigs (99); by contrast, three consecutive passages of the human A/Anhui/1/2013 (H7N9) virus in pigs did not increase virus titers or the preference for human-type receptors (99). These findings demonstrate that H7 influenza viruses already have the capability to transmit among mammals, which demonstrates their pandemic potential; even so, only two family clusters of human-to-human transmission of H7N9 viruses have been reported to date (107, 108). These studies also suggest that H7N9 virus passages in mammals do not readily result in highly virulent and/or mammalian-transmissible viruses.
In another study, a highly pathogenic H7N1 virus (A/ostrich/Italy/2332/2000) underwent 10 serial passages in ferrets (109). The wild-type isolate possessed avian-type receptor-binding specificity (88) but encoded the mammalian-adapting PB2-627K mutation (see ‘The Role of Polymerase Proteins in Influenza A Virus Transmission’). Serial passages in ferrets resulted in the selection of a variant that transmitted to other ferrets via contact and respiratory droplet transmission (109). The transmissible variant possessed five additional amino acid changes in its PB2 (T81I), NP (V284M), M1 (R95K, Q211K), and HA (K/R304R) proteins. The mutation in HA localizes to the stalk region and may affect the stability of the protein (see ‘The Role of HA Stability in Influenza A Virus Transmission).
Receptor-binding specificity and transmissibility of H5 influenza viruses
Most highly pathogenic avian H5N1 viruses, including those isolated from humans, bind to Siaα2,3Gal, but not to Siaα2,6Gal ((110); reviewed in (111)); this binding preference is consistent with the structural analysis of an H5 HA (112). The avian-type receptor-binding specificity of H5 HAs may explain why these viruses have not acquired the ability to transmit among humans. However, several H5N1 viruses isolated from infected people (113-115) and recent avian isolates from Egypt (116) bind somewhat to Siaα2,6Gal, while maintaining their ability to binding to Siaα2,3Gal. Several natural or experimentally introduced amino acid changes in HA (namely, N186K, E190D, K193R, Q226L, S227N, and G228S) increased binding to Siaα2,6Gal or decreased binding to Siaα2,3Gal (112, 113, 116-120). Another study revealed that mutations that affected the receptor-binding specificity of H1 HA (i.e., those at positions 190 and 225) did not confer human-type receptor-specificity to H5 HA; in contrast, mutations at positions 226 and 288 (which affect H3 HA receptor-binding specificity) resulted in an H5 HA that bound to a human-type receptor-analog (112).
Several studies have tested the transmissibility of highly pathogenic avian H5N1 influenza viruses in animal models, including isolates obtained from humans. The human H5N1 isolate A/Vietnam/1203/2004 (VN1203) did not transmit among mice (an animal model in which influenza viruses typically do not transmit) in the same cage (121). Seroconversion or infection of ferret cage mates was reported for two different H5N1 viruses, one of which also showed limited binding to human-type receptors (A/Hong Kong/213/2003) (115); however, virus transmission via respiratory droplets was not assessed in this study. Studies with several H5N1 viruses have demonstrated a lack of respiratory droplet transmission among ferrets (122-125), although seroconversion was reported for two of three animals housed adjacent to ferrets inoculated with A/Hong Kong/486/1997 (122).
H5N1 influenza viruses reassort readily with human H3N2 or 2009 pandemic H1N1 viruses in experimental settings (126-128). Several studies have, therefore, tested the transmissibility in mammals of avian H5N1/human reassortant viruses (122, 128, 129). Maines et al. (122) found that a virus possessing the HA and NA genes of an efficiently transmitting human H3N2 virus and the remaining genes from an H5N1 virus did not efficiently transmit via respiratory droplets, although one contact animal sero-converted. Similarly, the six internal genes (or the three polymerase and NP genes) of the human H3N2 virus did not confer respiratory droplet transmission in ferrets in combination with the HA and NA genes (or HA, NA, M, and NS genes) of the H5N1 virus (122). The virus possessing the polymerase and NP vRNA segments from the human H3N2 virus and the HA, NA, M, and NS vRNA segments from the H5N1 virus was then passaged five time in ferrets; yet, virus transmission among ferrets was still not detected (122). Likewise, none of five avian H5N1/human H3N2 virus reassortants (all possessing the H5 HA) transmitted to ferrets housed in the same cage (129). Schrauwen et al. (130) infected ferrets with a mixture of reassortant H5N1/2009 pandemic H1N1 viruses (all possessing the H5 HA gene) and found a dominant population of reassortants that carried the NA and M genes of 2009 pandemic H1N1 viruses; no respiratory droplet transmission was detected for these viruses (note, this finding may indicate a dominance of the 2009 pandemic H1N1 NA and M vRNA segments over the NA and M vRNA segments of previously circulating viruses). Likewise, a reassortant virus possessing an H5 HA in the background of a 2009 pandemic H1N1 virus did not transmit to contact ferrets (131).
To determine whether the introduction of human-type amino acids into the HA of H5N1 viruses confers respiratory droplet transmissibility to these viruses in mammals, Maines et al. (118) generated several H5N1 viruses with mutations known to affect receptor-binding specificity (including HA-E190D, -K193S, -Q226L, and/or -G228S). Two mutants (encoding HA-226L/228S or HA-187G/190D/193S/226L/228S) were able to recognize Siaα2,6Gal in addition to Siaα2,3Gal (118). One mutant (possessing the HA-R193K, -Q226L, and -G228S mutations) exhibited a shift from avian- to human-type receptor-binding specificity; however, this shift did not result in virus transmission via respiratory droplets in the ferret model (118). In another study, Chen et al. (125) demonstrated that three mutations in HA (HA-Q196R, -Q226L, -G228S) conferred predominant binding to Siaα2,6Gal; this mutant H5N1 virus was transmitted to co-housed ferrets but not to animals placed in a separate cage. However, a reassortant in which the avian H5N1-derived N1 gene was replaced with an N2 gene of human virus origin transmitted via respiratory droplets to one of two ferrets housed in a separate cage; the infected ferret did not succumb to the infection (125). This was the first study that resulted in a respiratory droplet-transmissible virus possessing an H5 HA gene derived from a highly pathogenic avian H5N1 influenza virus.
Fouchier and colleagues (123) tested an H5N1 virus encoding the HA-Q226L and -G228S mutations (known to confer efficient binding to human-type receptors), and the PB2-R627K mutation, which is known to confer efficient replication in mammals (see ‘The Role of Polymerase Proteins in Influenza A Virus Transmission’); virus transmission via respiratory droplets did not occur. However, after ten consecutive passages of this virus in ferrets (in which nasal turbinate or nasal wash samples obtained from infected animals were used to inoculate the next set of ferrets), a variant was isolated that transmitted via respiratory droplets among ferrets; this virus was not lethal in ferrets. Three mutations (HA-H110Y and -T160A; PB1-H99Y), in addition to those introduced deliberately, were essential for the respiratory droplet transmissibility of this virus among ferrets (123, 132).
By using an approach in which ‘virus libraries’ comprising a large number of mutants that possessed random amino acid changes in the globular head of their H5 HA were screened for variants that efficiently bound to human-type receptors, we isolated a mutant (possessing the HA-N224K and -Q226L mutations) whose receptor-binding specificity shifted from Siaα2,3Gal to Siaα2,6Gal binding (124). A reassortant possessing the mutant HA together with the remaining seven vRNA segments of A/California/04/2009 (a prototype human 2009 pandemic H1N1 virus) did not transmit to ferrets housed in adjacent cages, but gained this function after two passages in ferrets (124); all infected animals recovered from the infection. Two additional mutations (HA-N158D and -T318I) were detected in the HA of this ferret-transmissible virus (124).
Structural studies demonstrated that the mutations in the HA proteins of the ferret-transmissible viruses isolated by Herfst et al. (123) and Imai et al. (124) allowed H5 HA binding to Siaα2,6Gal in a conformation similar to that observed for human H1, H2, and H3 viruses (96, 133, 134). In particular, Xiong et al. (96) found that the orientation of galactose and N-acetylglucosamine at positions 2 and 3 of the sialic acid resemble those found for human viruses that interact with human-type receptors; this orientation differs from that of wild-type H5N1 HA interacting with a human-type receptor. Studies in guinea pigs have shown that several H5N1 viruses can transmit to co-housed animals (135, 136); the mutations HA-160A (which results in the loss of a glycosylation site at positions 158–160 of HA; see ‘The Role of HA Glycosylation in Influenza A Virus Transmission’) and PB2-627K or PB2-701N (see ‘The Role of Polymerase Proteins in Influenza A Virus Transmission’) are responsible for this feature. A study that tested reassortants between a highly pathogenic avian H5N1 virus (with dual avian/human receptor-binding specificity) and a human 2009 pandemic H1N1 virus found that the human virus PA or NS gene conferred respiratory droplet transmissibility to the H5N1 virus in guinea pigs (128); in addition, the NA and M genes of the human virus contributed to the respiratory droplet transmission in this animal model (128).
Receptor-binding specificity and transmissibility of H9 influenza viruses
Human infections with H9 influenza viruses have been infrequently reported (4, 137, 138), but epidemiological studies suggest that they may occur more frequently than currently recognized (reviewed in (139)). Viruses of the H9 subtype circulate widely in poultry and have also been isolated from pigs. Importantly, H9 viruses have donated genes to the currently circulating H5N1 (140-146) and H7N9 viruses (see ‘Sporadic Infections of Humans with Swine or Avian Influenza Viruses’). Many Asian H9N2 viruses have acquired the HA-Q226L mutation, which increases binding to human-type receptors (147, 148). Nonetheless, several avian H9N2 viruses tested did not transmit among ferrets via respiratory droplets, although two of these viruses infected co-housed, naïve animals (149). A reassortant possessing Asian H9N2 virus-derived HA (encoding HA-226L) and NA genes combined with the internal genes of a human H3N2 virus transmitted to ferret cage mates, but not via respiratory droplets to animals in adjacent cages (149). However, after ten sequential passages in ferrets, a virus was obtained that transmitted via respiratory droplets (150). Three amino acid changes in the surface glycoproteins (HA-T189A and -G510R, and NA-I28V) were responsible for this change in transmissibility (150). Based on this information, Kimble et al. (151) generated reassortants composed of 2009 pandemic H1N1 vRNA segments and the HA or HA/NA vRNA segments of the H9N2 ferret-transmissible virus, or the parent of the ferret-transmissible virus. Three of four reassortant viruses transmitted via respiratory droplets to naïve ferrets (151), further demonstrating the potential of avian/human reassortants to transmit among mammals via respiratory droplets.
Receptor-binding specificity and transmissibility of H6 influenza viruses
Only few infections of humans with influenza viruses of the H6 subtype have been reported (reviewed in (146)); however, the recent testing of 257 H6 viruses isolated in China from 2008–2011 revealed that 87 of them bound to human-type receptors (152). Many of these viruses replicated efficiently in mice and five of ten viruses tested transmitted to co-housed guinea pigs (152). Avian H6 viruses may thus be capable of transmitting and adapting to humans.
Collectively, receptor-binding and transmission studies of H1, H5, H6, H7, and H9 viruses established that HA receptor-binding specificity plays a pivotal role in virus transmissibility. HA proteins with avian-type receptor preference do not facilitate virus transmission among ferrets. Viruses that bind to both avian- and human-type cellular receptors may transmit among ferrets via respiratory droplets, but the efficiency of transmission will likely be low. A complete change from Siaα2,3Gal to Siaα2,6Gal binding may be necessary (although not sufficient) to allow efficient respiratory droplet transmission in ferrets, and perhaps humans. This change in receptor-binding specificity may be important to avoid virus binding to mucus in the respiratory tract of humans, which predominantly contains Siaα2,3Gal (47, 153).
The Role of HA Glycosylation in Influenza A Virus Transmission
Newly synthesized HA protein is inserted into the ER and transported through the Golgi complex to the plasma membrane. During its transport through the ER/Golgi network, HA is glycosylated at asparagine residues of the N-x-S/T motif (N-glycosylation). The number and location of glycosylation sites and oligosaccharide side chains differ among influenza strains and subtypes (reviewed in (154)). HA glycosylation affects a variety of biological properties including antigenicity, virulence, receptor-binding specificity, and receptor-binding affinity (reviewed in (154)). Glycosylation had not been linked to influenza virus transmissibility until two key observations were made: both ferret-transmissible H5 viruses isolated by Imai et al. (124) and Herfst et al. (123) possessed mutations (HA-N158D and HA-T160A, respectively) that resulted in the loss of the same glycosylation site (i.e., the N-x-S/T glycosylation site at position 158–160 of HA). Moreover, the HAs of the ferret- and guinea pig-transmissible H5 viruses described by Chen et al. (125) and Zhang et al. (128) also lack a glycosylation site at position 158–160 of HA. Thus, all four mammalian-transmissible H5 viruses described to date lack this glycosylation site, suggesting that glycosylation at this site may sterically interfere with virus binding to the cellular receptor. This idea is supported by the findings that the loss of the glycosylation site at HA position 158–160 results in increased binding of H5N1 viruses to human-type receptors (136, 155), and in efficient replication in the upper respiratory tract of ferrets of H5N1 viruses possessing the HA-Q226L or –Q226L/G228S mutations (155). Our analysis of H5N1 virus sequences revealed that H5N1 viruses isolated from humans in Egypt lack the glycosylation site at HA 158–160, whereas one-fourth of Egyptian avian H5N1 viruses encode a glycosylation site at this position (156); this finding may suggest that H5N1 viruses without the glycosylation site transmit more efficiently to humans than do those with a glycosylation site at HA 158–160. In line with the hypothesis, it is noteworthy that the HA proteins of the recently emerged H7N9 viruses naturally lack the glycosylation site at HA 158–160, which may contribute to their ability to infect humans.
The Role of HA Stability in Influenza A Virus Transmission
The ferret-transmissible virus isolated by Imai et al. (124) acquired an HA-T318I mutation during passage in ferrets. The localization of this mutation in the stem region of HA suggested a potential effect on HA stability which we, indeed, demonstrated (124). The heat stability of HA is also associated with the pH at which HA undergoes a conformational change that triggers the fusion of the viral and endosomal membranes: in influenza virus-infected cells, the low pH in late endosomes triggers the conformational change in HA, but the energy barrier for the conformational change can also be overcome by increased temperatures.
The significance of our finding that HA stability plays a role in virus transmission became evident when we found that the HA-N224K/Q226L mutations (which cause a change from human- to avian-type receptor-binding specificity; see ‘Receptor-binding specificity and transmissibility of H5 influenza viruses’) reduced heat stability, which was restored by the HA-T318I mutation (124). Even more interestingly, the HA-H110Y mutation detected in the ferret-transmissible virus isolated by Herfst et al. (123) was also shown recently to increase heat stability (132, 157). Together, these findings suggest that the acquisition of human-type receptor-binding specificity results in an unfavorable change in HA stability that requires compensatory mutations to offset the reduction in virus fitness. Two independent studies thus found the same mechanistic explanation for amino acid changes in HA, indicating that receptor-binding specificity and HA stability/fusion are mechanistically linked to determine virus transmissibility.
To date, more than 70 mutations in H1, H2, H3, H5, and H7 HA proteins have been described that affect HA heat stability and the pH at which fusion occurs (reviewed in (158)). The K387I mutation in an H5 HA reduced the pH value at which the conformational change occurred (159) and attenuated H5N1 virus replication in ducks (160). However, this mutation enhanced virus replication in mice (161) and in the upper respiratory tract of ferrets (162). Another study demonstrated that higher pH optima for the HA conformational change (in the range of pH 5.2–6.0) correlated with increased virulence in avian species (163).
Collectively, these findings established that avian and human influenza viruses have different pH optima for fusion, and that adapting changes which affect HA stability and the pH optimum for fusion may be critical to facilitate virus transmission and the establishment of new lineages.
The Role of NA in Influenza A Virus Transmission
The NA protein encodes a sialidase that cleaves sialic acids from sialyloligosaccharides; this enzymatic activity is vital for efficient infection and release of viruses from host cells, as the virus would otherwise be trapped by sialic acids on mucus or remain attached to the sialic acids on the host cells, resulting in large viral aggregates. HA-mediated binding to, and NA-mediated release from sialic acids therefore must be balanced for efficient virus replication. The NA proteins of all influenza viruses exhibit significantly higher enzymatic activity against Siaα2,3Gal than against Siaα2,6Gal (164-166); nonetheless, the N1 NA of a 2009 pandemic virus had detectable activity against Siaα2,6Gal (166), similar to NA proteins derived from human N2 viruses (164, 165). Interestingly, several of the recently identified respiratory droplet-transmissible H5 viruses possess an NA gene derived from a human N2 virus Chen et al. (125) or from a 2009 pandemic H1N1 virus (124, 128). Replacement of the N2 NA gene of an H1N2 swine virus (which differed from the 2009 pandemic H1N1 virus by only the NA vRNA segment) with the 2009 pandemic H1N1 NA gene increased virus transmission by respiratory droplet transmission in ferrets (167). In contrast, Zhang et al. (128) and Herfst et al. (123) identified respiratory droplet-transmissible H5 viruses possessing the avian virus NA vRNA segment, demonstrating that a human virus NA vRNA segment is not essential for respiratory droplet transmissibility in mammals.
NA proteins may possess in-frame deletions that result in ‘stalks’ of different lengths (the stalk anchors the NA globular head, including the enzymatic center, to the virus membrane). In particular, a 20-amino acid deletion has been prevalent in the stalk of H5N1 virus NA proteins since 2003. The length of the NA stalk affects influenza virulence (168, 169) and may facilitate the adaptation of avian influenza viruses to terrestrial poultry (142, 170-174). Two 2009 pandemic H1N1 viruses encoding H5N1 virus-derived NA proteins with long or short stalks transmitted to ferrets in the same cage, but only the virus encoding the NA with the long stalk also transmitted via respiratory droplets (175). The transmissible virus had higher neuraminidase activity (175), which may have facilitated its release from host cells and its transmission to ferrets housed in adjacent cages. These findings suggest that the length of the NA stalk may contribute to influenza virus transmission.
The Role of Polymerase Proteins in Influenza A Virus Transmission
Influenza viral replication and transcription are catalyzed by the viral polymerase complex, composed of the PB2, PB1, and PA proteins. Although all three polymerase subunits affect influenza virulence, PB2 is the main polymerase determinant for influenza virulence and host range and thus, for influenza virus adaptation to new hosts. Studies by Subbarao et al. (176) and Hatta et al. (177) found that the amino acid at position 627 of PB2 is critical for efficient influenza virus replication in mammalian cells. Most human influenza viruses (with the exception of 2009 pandemic H1N1 viruses, see below) encode a lysine at this position, which confers efficient viral replication in the upper respiratory tract of mammals (178). A glutamic acid residue at PB2-627 (encoded by most avian virus PB2 proteins) restricts virus replication in mammals, particularly at 33°C (i.e., the temperature of the upper respiratory tract) (178). The PB2 proteins of the 1918, 1957, 1968, and 1977 pandemic viruses are from the same PB2 ‘lineage’ and all encode PB2-627K. In 2005, highly pathogenic avian H5N1 viruses caused an outbreak among several avian species at Qinghai Lake, China (179-181). In some of these viruses, the PB2-627K mutation emerged, and descendants of these viruses have spread westward into Europe and the Middle East, so that all contemporary Middle Eastern H5N1 viruses encode the mammalian-adapting PB2-627K residue. Moreover, PB2-627K is frequently selected during the replication of avian influenza viruses not only in humans and other mammals, but also in poultry (reviewed in (154)). Together, these findings established the significance of PB2-627K for influenza virus adaptation to new hosts.
The 2009 pandemic H1N1 PB2 proteins encode PB2-627E (i.e., the avian-type amino acid). However, a basic residue at PB2 position 591 (in the three-dimensional structure located close to position 627) was shown to compensate for the lack of PB2-627K (182, 183). Other residues in PB2 also affect virulence and host range. An aspartic acid (encoded by most avian influenza viruses) to asparagine change at position 701 increased the virulence of avian influenza viruses in mammals (184, 185). The amino acid at position PB2-701 interacts with the cellular nuclear import factor importin α; asparagine facilitates a stronger interaction with importin α in mammalian cells relative to avian cells, resulting in increased replicative ability (186). The aspartic acid-to-asparagine change has been detected upon replication of avian influenza viruses in humans, other mammals, and ostriches (see Influenza Research Database at www.fludb.org), underscoring the role of this mutation for influenza virus adaptation to novel hosts. In addition, the amino acid at position 271 of PB2 affects the replicative ability of influenza viruses: PB2-271A (found in most human influenza viruses) confers higher replication in mammalian cells and mice than does PB2-271T (typically found in avian influenza viruses) (187).
The role of PB2 in influenza virus transmissibility was established in several studies: one experiment demonstrated that the HA and NA genes of the 1918 pandemic virus did not confer respiratory droplet transmission to an avian influenza virus in ferrets, but the addition of the 1918 pandemic virus PB2 gene conferred this ability (63). Specially, the human-type PB2-627K and/or -701N residues conferred higher virus transmissibility in guinea pigs in the genetic background of an H5N1 (135, 136) or human H3N2 (135) viruses. Another study found that an HA-Q226R or a PB2-A271T mutation in a 2009 pandemic H1N1 virus abolished respiratory droplet transmission in guinea pigs, and that both mutations together abolished respiratory droplet transmission in ferrets (188). The partially transmissible H5 virus described by Chen et al. (125) did not encode a human-type amino acid at PB2-627, -701, -or 271, suggesting that limited H5 virus transmission in guinea pigs can be achieved without these amino acid markers in PB2. By contrast, the ferret-transmissible H5 viruses isolated by Herfst et al. (123) and Imai et al. (124) possessed the intentionally introduced PB2-627K mutation or the PB2 gene of a 2009 pandemic H1N1 virus (hence encoding a basic residue at position 591), respectively. In another study, Zhang et al. reported that the PA or NS vRNA segment of a human virus was sufficient to render an avian H5N1 virus respiratory droplet transmissible in guinea pigs (128); interestingly, the avian H5N1 virus used in this study (A/duck/Guanxi/35/2001) encodes the human-adapting PB1-701N residue.
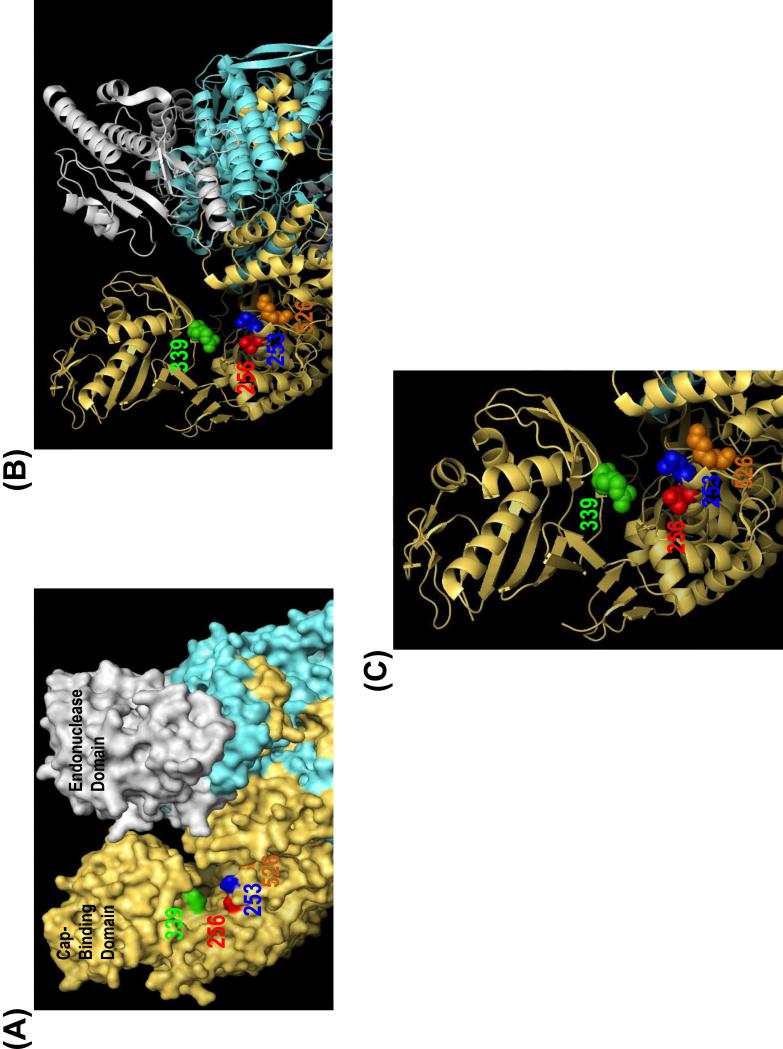
The recent publications of the X-ray crystallographic structures of influenza A and B virus polymerase complexes (189, 190) allow this structural information to be correlated with amino acid residues that affect replicative ability, virulence, and host range. We mapped mutations known to affect influenza viral replicative ability onto the structure and, in addition to the known ‘627 domain’, identified an area in which several of the amino acids that affect replication are located (Figure): Position 339, which is involved in cap-binding (191) and affects virulence (191-193); position 253, which affects the polymerase activity of an H9 virus polymerase complex (194); position 256, which affects the polymerase activity of an avian H5N1 influenza virus (195); and position 526 which, together with other positions in PB2, affects H5N1 viral polymerase activity (196). Based on their location, the amino acid residues at these positions may interact with a common factor, such as the cap structure of mRNAs or a host protein.
Figure. Location of the PB2 residues 253, 256, 339, and 526, which affect viral replication.
Shown is the three-dimensional structure of an influenza A virus polymerase complex (Protein Data Bank ID 4WSB). The PB2, PB1, and PA subunits are shown in gold, blue, and gray, respectively. PB2 residues 253, 256, 339, and 526 are indicated in blue, red, green, and orange, respectively. (A) Surface view. (B) ‘Ribbon view’ to depict α-helices and β-sheets. Amino acid spheres are shown for PB2 residues 253, 256, 339, and 526. (C) Enlarged view of (B).
The Role of the ‘Triple Reassortant Internal Gene’ Cassette in Influenza A Virus Transmission
Viruses with the ‘triple reassortant internal genes’ that emerged in North American swine viruses in 1998 replaced the classical H1N1 swine viruses in North America that had circulated for six decades, attesting to the ability of viruses with these genes to establish a new lineage, and to replace the previously circulating one. As briefly described in the section on ‘Establishment of Novel Influenza A Virus Lineages in Humans’, these viruses possess the following segments: PB2 and PA polymerase vRNA segments derived from avian influenza viruses; nucleoprotein (NP), matrix (M), and nonstructural (NS) vRNA segments derived from previously circulating H1N1 swine influenza viruses; and HA, NA, and PB1 vRNA segments derived from human influenza viruses (13-16). The transmissibility of these viruses in ferrets was found to depend primarily on the origin of the HA and NA genes (197-199); however, the polymerase PB2, PB1, and PA genes also affected transmissibility (199).
The 2009 pandemic H1N1 viruses differ from some of the North American triple reassortant swine viruses in the origin of their NA and M vRNA segments (see ‘Establishment of Novel Influenza Virus Lineages in Humans’). While 2009 pandemic H1N1 viruses transmit efficiently among ferrets via respiratory droplets (reviewed in (21, 154)), a reassortant possessing the NA and M vRNA segments of a North American triple reassortant swine virus isolated from an infected person (A/Ohio/02/2007) did not transmit to all exposed ferrets (200). This finding suggested that the Eurasian avian-like swine virus NA and M vRNA segments of 2009 pandemic H1N1 viruses may contribute to the transmissibility of these viruses, at least in the ferret model. Other studies demonstrated that A/Puerto Rico/8/34 (H1N1, a strain commonly used in influenza virus research) does not transmit via respiratory droplets among guinea pigs; however, PR8 reassortant viruses possessing the 2009 pandemic H1N1 HA, NA, and M vRNA segments (201) or the 2009 pandemic H1N1 M vRNA segment (202) were transmitted among subsets of guinea pigs via direct contact or respiratory droplets, respectively. Likewise, the 2009 pandemic H1N1 M vRNA segment increased the transmissibility of a triple reassortant swine virus that possessed the 2009 pandemic H1N1 HA and NA genes (202). These finding are consistent with a study that demonstrated the importance of the 2009 pandemic H1N1 HA and M genes for virus replication and transmissibility in pigs (203). Collectively, these studies suggest that the 2009 pandemic H1N1 NA and M vRNA segments are important for the transmissibility of 2009 pandemic H1N1 viruses. The importance of the NA segments may stem from its ability to confer relatively high neuraminidase activity and efficient particle release from infected cells (200, 201).
Conclusion
Recent studies have demonstrated that avian influenza viruses of different subtypes may acquire the ability to transmit among mammals. Several features including HA receptor-binding specificity, HA stability, HA glycosylation, and viral replicative ability in mammalian cells likely act together to allow avian influenza viruses to adapt to, replicate in, and transmit among mammals.
Highlights.
HA receptor-binding specificity is important for virus transmissibility
The polymerase complex is also important for virus transmissibility
Avian influenza viruses may acquire the ability to transmit among mammals
Acknowledgements
We thank Sue Watson for editing the manuscript. Funding support came from the Center for Research on Influenza Pathogenesis (CRIP) funded by the NIAID Contract HHSN272201400008C; the Japan Initiative for Global Research Network on Infectious Diseases; Grants-in-Aid for Specially Promoted Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan; Grants-in-Aid from the Ministry of Health, Labour and Welfare, Japan; and from Strategic Basic Research Programs of the Japanese Science and Technology Agency.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- 1.Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. A distinct lineage of influenza A virus from bats. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arzey GG, Kirkland PD, Arzey KE, Frost M, Maywood P, Conaty S, Hurt AC, Deng YM, lannello P, Barr I, Dwyer DE, Ratnamohan M, McPhie K, Selleck P. Influenza virus A (H10N7) in chickens and poultry abattoir workers, Australia. Emerg Infect Dis. 2012;18:814–816. doi: 10.3201/eid1805.111852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, Orr WK, Shortridge KF. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 5.Yuan J, Zhang L, Kan X, Jiang L, Yang J, Guo Z, Ren Q. Origin and molecular characteristics of a novel 2013 avian influenza A(H6N1) virus causing human infection in Taiwan. Clin Infect Dis. 2013;57:1367–1368. doi: 10.1093/cid/cit479. [DOI] [PubMed] [Google Scholar]
- 6.Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, Fan G, Yang F, Li X, Zhou J, Zou S, Yang L, Chen T, Dong L, Bo H, Zhao X, Zhang Y, Lan Y, Bai T, Dong J, Li Q, Wang S, Zhang Y, Li H, Gong T, Shi Y, Ni X, Li J, Zhou J, Fan J, Wu J, Zhou X, Hu M, Wan J, Yang W, Li D, Wu G, Feng Z, Gao GF, Wang Y, Jin Q, Liu M, Shu Y. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet. 2014;383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- 7.Ostrowsky B, Huang A, Terry W, Anton D, Brunagel B, Traynor L, Abid S, Johnson G, Kacica M, Katz J, Edwards L, Lindstrom S, Klimov A, Uyeki TM. Low pathogenic avian influenza A (H7N2) virus infection in immunocompromised adult, New York, USA, 2003. Emerg Infect Dis. 2012;18:1128–1131. doi: 10.3201/eid1807.111913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez-Martinez I, Balish A, Barrera-Badillo G, Jones J, Nunez-Garcia TE, Jang Y, Aparicio-Antonio R, Azziz-Baumgartner E, Belser JA, Ramirez-Gonzalez JE, Pedersen JC, Ortiz-Alcantara J, Gonzalez-Duran E, Shu B, Emery SL, Poh MK, Reyes-Teran G, Vazquez-Perez JA, Avila-Rios S, Uyeki T, Lindstrom S, Villanueva J, Tokars J, Ruiz-Matus C, Gonzalez-Roldan JF, Schmitt B, Klimov A, Cox N, Kuri-Morales P, Davis CT, Diaz-Quinonez JA. Highly pathogenic avian influenza A(H7N3) virus in poultry workers, Mexico, 2012. Emerg Infect Dis. 2013;19:1531–1534. doi: 10.3201/eid1909.130087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rhmmerzwaan GF, Schutten M, Van Doornum GJ, Koch G, Bosman A, Koopmans M, Osterhaus AD. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A. 2004;101:1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koopmans M, Wilbrink B, Conyn M, Natrop G, van der Nat H, Vennema H, Meijer A, van Steenbergen J, Fouchier R, Osterhaus A, Bosman A. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet. 2004;363:587–593. doi: 10.1016/S0140-6736(04)15589-X. [DOI] [PubMed] [Google Scholar]
- 11.Shaw ML, Palese P. Orthomyxoviridae. In: Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B, editors. Fields Virology. Sixth ed One. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2013. pp. 1151–1185. [Google Scholar]
- 12.Taubenberger JK, Baltimore D, Doherty PC, Markel H, Morens DM, Webster RG, Wilson IA. Reconstruction of the 1918 influenza virus: unexpected rewards from the past. MBio. 2012;3 doi: 10.1128/mBio.00201-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karasin AI, Schutten MM, Cooper LA, Smith CB, Subbarao K, Anderson GA, Carman S, Olsen CW. Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977-1999: evidence for wholly human and reassortant virus genotypes. Virus Res. 2000;68:71–85. doi: 10.1016/s0168-1702(00)00154-4. [DOI] [PubMed] [Google Scholar]
- 14.Olsen CW. The emergence of novel swine influenza viruses in North America. Virus Res. 2002;85:199–210. doi: 10.1016/s0168-1702(02)00027-8. [DOI] [PubMed] [Google Scholar]
- 15.Webby RJ, Swenson SL, Krauss SL, Gerrish PJ, Goyal SM, Webster RG. Evolution of swine H3N2 influenza viruses in the United States. J Virol. 2000;74:8243–8251. doi: 10.1128/jvi.74.18.8243-8251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon K, Krauss S, Webster RG. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol. 1999;73:8851–8856. doi: 10.1128/jvi.73.10.8851-8856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bastien N, Antonishyn NA, Brandt K, Wong CE, Chokani K, Vegh N, Horsman GB, Tyler S, Graham MR, Plummer FA, Levett PN, Li Y. Human infection with triple-reassortant swine influenza A(H1N1) virus containing the hemagglutinin and neuraminidase genes of seasonal influenza virus. J Infect Dis. 2010;201:1178–1182. doi: 10.1086/651507. [DOI] [PubMed] [Google Scholar]
- 18.Cox CM, Neises D, Garten RJ, Bryant B, Hesse RA, Anderson GA, Trevino-Garrison I, Shu B, Lindstrom S, Klimov AI, Finelli L. Swine influenza virus A (H3N2) infection in human, Kansas, USA, 2009. Emerg Infect Dis. 2011;17:1143–1144. doi: 10.3201/eid1706.101488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olsen CW, Karasin AI, Carman S, Li Y, Bastien N, Ojkic D, Alves D, Charbonneau G, Henning BM, Low DE, Burton L, Broukhanski G. Triple reassortant H3N2 influenza A viruses, Canada, 2005. Emerg Infect Dis. 2006;12:1132–1135. doi: 10.3201/eid1207.060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shinde V, Bridges CB, Uyeki TM, Shu B, Balish A, Xu X, Lindstrom S, Gubareva LV, Deyde V, Garten RJ, Harris M, Gerber S, Vagasky S, Smith F, Pascoe N, Martin K, Dufficy D, Ritger K, Conover C, Quinlisk P, Klimov A, Bresee JS, Finelli L. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005-2009. N Engl J Med. 2009;360:2616–2625. doi: 10.1056/NEJMoa0903812. [DOI] [PubMed] [Google Scholar]
- 21.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaydos JC, Hodder RA, Top FH, Jr., Soden VJ, Allen RG, Bartley JD, Zabkar JH, Nowosiwsky T, Russell PK. Swine influenza A at Fort Dix, New Jersey (January-February 1976). I. Case finding and clinical study of cases. J Infect Dis. 1977;136(Suppl):S356–362. doi: 10.1093/infdis/136.supplement_3.s356. [DOI] [PubMed] [Google Scholar]
- 23.Goldfield M, Bartley JD, Pizzuti W, Black HC, Altman R, Halperin WE. Influenza in New Jersey in 1976: isolations of influenza A/New Jersey/76 virus at Fort Dix. J Infect Dis. 1977;136(Suppl):S347–355. doi: 10.1093/infdis/136.supplement_3.s347. [DOI] [PubMed] [Google Scholar]
- 24.Hodder RA, Gaydos JC, Allen RG, Top FH, Jr., Nowosiwsky T, Russell PK. Swine influenza A at Fort Dix, New Jersey (January-February 1976). III. Extent of spread and duration of the outbreak. J Infect Dis. 1977;136(Suppl):S369–375. doi: 10.1093/infdis/136.supplement_3.s369. [DOI] [PubMed] [Google Scholar]
- 25.Kendal AP, Goldfield M, Noble GR, Dowdle WR. Identification and preliminary antigenic analysis of swine influenza-like viruses isolated during an influenza outbreak at Fort Dix, New Jersey. J Infect Dis. 1977;136(Suppl):S381–385. doi: 10.1093/infdis/136.supplement_3.s381. [DOI] [PubMed] [Google Scholar]
- 26.Dowdle WR, Millar JD. Swine influenza: lessons learned. Med Clin North Am. 1978;62:10471057. doi: 10.1016/s0025-7125(16)31754-0. [DOI] [PubMed] [Google Scholar]
- 27.Kitikoon P, Vincent AL, Gauger PC, Schlink SN, Bayles DO, Gramer MR, Darnell D, Webby RJ, Lager KM, Swenson SL, Klimov A. Pathogenicity and transmission in pigs of the novel A(H3N2)v influenza virus isolated from humans and characterization of swine H3N2 viruses isolated in 2010-2011. J Virol. 2012;86:6804–6814. doi: 10.1128/JVI.00197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson MI, Vincent AL, Kitikoon P, Holmes EC, Gramer MR. Evolution of novel reassortant A/H3N2 influenza viruses in North American swine and humans, 2009-2011. J Virol. 2012;86:8872–8878. doi: 10.1128/JVI.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ducatez MF, Hause B, Stigger-Rosser E, Darnell D, Corzo C, Juleen K, Simonson R, Brockwell-Staats C, Rubrum A, Wang D, Webb A, Crumpton JC, Lowe J, Gramer M, Webby RJ. Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States. Emerg Infect Dis. 2011;17:1624–1629. doi: 10.3201/1709.110338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Q, Ma J, Liu H, Qi W, Anderson J, Henry SC, Hesse RA, Richt JA, Ma W. Emergence of novel reassortant H3N2 swine influenza viruses with the 2009 pandemic H1N1 genes in the United States. Arch Virol. 2012;157:555–562. doi: 10.1007/s00705-011-1203-9. [DOI] [PubMed] [Google Scholar]
- 31.Claas EC, de Jong JC, van Beek R, Rimmelzwaan GF, Osterhaus AD. Human influenza virus A/HongKong/156/97 (H5N1) infection. Vaccine. 1998;16:977–978. doi: 10.1016/s0264-410x(98)00005-x. [DOI] [PubMed] [Google Scholar]
- 32.Claas EC, Osterhaus AD, van Beek R, De Jong JC, Rimmelzwaan GF, Senne DA, Krauss S, Shortridge KF, Webster RG. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 33.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 34.Belser JA, Bridges CB, Katz JM, Tumpey TM. Past, present, and possible future human infection with influenza virus A subtype H7. Emerg Infect Dis. 2009;15:859–865. doi: 10.3201/eid1506.090072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.China—WHO Joint Mission on Human Infection with Avian Influenza A(H7N9) Virus. 2013 http://www.who.int/influenza/human_animal_interface/influenza_h7n9/ChinaH7N9JointMissionReport2013.pdf.
- 36.Centers for Disease C, Prevention Emergence of Avian Influenza A(H7N9) Virus Causing Severe Human Illness - China, February-April 2013. MMWR Morb Mortal Wkly Rep. 2013;62:366–371. [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y, Liang W, Yang S, Wu N, Gao H, Sheng J, Yao H, Wo J, Fang Q, Cui D, Li Y, Yao X, Zhang Y, Wu H, Zheng S, Diao H, Xia S, Zhang Y, Chan KH, Tsoi HW, Teng JL, Song W, Wang P, Lau SY, Zheng M, Chan JF, To KK, Chen H, Li L, Yuen KY. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet. 2013 doi: 10.1016/S0140-6736(13)60903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 39.Kageyama T, Fujisaki S, Takashita E, Xu H, Yamada S, Uchida Y, Neumann G, Saito T, Kawaoka Y, Tashiro M. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. 2013;18:20453. [PMC free article] [PubMed] [Google Scholar]
- 40.Lam TT, Wang J, Shen Y, Zhou B, Duan L, Cheung CL, Ma C, Lycett SJ, Leung CY, Chen X, Li L, Hong W, Chai Y, Zhou L, Liang H, Ou Z, Liu Y, Farooqui A, Kelvin DJ, Poon LL, Smith DK, Pybus OG, Leung GM, Shu Y, Webster RG, Webby RJ, Peiris JS, Rambaut A, Zhu H, Guan Y. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature. 2013 doi: 10.1038/nature12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J, Liu W, Zhao G, Yang W, Wang Y, Ma J, Shu Y, Lei F, Gao GF. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013;381:1926–1932. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- 42.Shi JZ, Deng GH, Liu PH, Zhou JP, Guan LZ, Li WH, Li XY, Guo J, Wang GJ, Fan J, Wang JL, Li YY, Jiang YP, Liu LL, Tian GB, Li CJ, Chen HL. Isolation and characterization of H7N9 viruses from live poultry markets - Implication of the source of current H7N9 infection in humans. Chinese Science Bulletin. 2013;58:1857–1863. [Google Scholar]
- 43.Wu A, Su C, Wang D, Peng Y, Liu M, Hua S, Li T, Gao GF, Tang H, Chen J, Liu X, Shu Y, Peng D, Jiang T. Sequential Reassortments Underlie Diverse Influenza H7N9 Genotypes in China. Cell Host Microbe. 2013 doi: 10.1016/j.chom.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 44.Ito T, Kawaoka Y. Host-range barrier of influenza A viruses. Vet Microbiol. 2000;74:71–75. doi: 10.1016/s0378-1135(00)00167-x. [DOI] [PubMed] [Google Scholar]
- 45.Rogers GN, Paulson JC. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983;127:361–373. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- 46.Rogers GN, Pritchett TJ, Lane JL, Paulson JC. Differential sensitivity of human, avian, and equine influenza A viruses to a glycoprotein inhibitor of infection: selection of receptor specific variants. Virology. 1983;131:394–408. doi: 10.1016/0042-6822(83)90507-x. [DOI] [PubMed] [Google Scholar]
- 47.Couceiro JN, Paulson JC, Baum LG. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 1993;29:155–165. doi: 10.1016/0168-1702(93)90056-s. [DOI] [PubMed] [Google Scholar]
- 48.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci U S A. 2004;101:4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 50.Baum LG, Paulson JC. Sialyloligosaccharides of the respiratory epithelium in the selection of human influenza virus receptor specificity. Acta Histochem Suppl. 1990;40:35–38. [PubMed] [Google Scholar]
- 51.Nicholls JM, Chan MC, Chan WY, Wong HK, Cheung CY, Kwong DL, Wong MP, Chui WH, Poon LL, Tsao SW, Guan Y, Peiris JS. Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nat Med. 2007;13:147–149. doi: 10.1038/nm1529. [DOI] [PubMed] [Google Scholar]
- 52.Yao L, Korteweg C, Hsueh W, Gu J. Avian influenza receptor expression in H5N1-infected and noninfected human tissues. FASEB J. 2008;22:733–740. doi: 10.1096/fj.06-7880com. [DOI] [PubMed] [Google Scholar]
- 53.van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. H5N1 Virus Attachment to Lower Respiratory Tract. Science. 2006;312:399. doi: 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- 54.Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000;74:8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, Donatelli I, Kida H, Paulson JC, Webster RG, Kawaoka Y. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nelli RK, Kuchipudi SV, White GA, Perez BB, Dunham SP, Chang KC. Comparative distribution of human and avian type sialic acid influenza receptors in the pig. BMC Vet Res. 2010;6:4. doi: 10.1186/1746-6148-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trebbien R, Larsen LE, Viuff BM. Distribution of sialic acid receptors and influenza A virus of avian and swine origin in experimentally infected pigs. Virol J. 2011;8:434. doi: 10.1186/1743-422X-8-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Costa T, Chaves AJ, Valle R, Darji A, van Riel D, Kuiken T, Majo N, Ramis A. Distribution patterns of influenza virus receptors and viral attachment patterns in the respiratory and intestinal tracts of seven avian species. Vet Res. 2012;43:28. doi: 10.1186/1297-9716-43-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Glaser L, Stevens J, Zamarin D, Wilson IA, Garcia-Sastre A, Tumpey TM, Basler CF, Taubenberger JK, Palese P. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J Virol. 2005;79:11533–11536. doi: 10.1128/JVI.79.17.11533-11536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burke DF, Smith DJ. A Recommended Numbering Scheme for Influenza A HA Subtypes. PLoSOne. 2014;9:e112302. doi: 10.1371/journal.pone.0112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tumpey TM, Maines TR, Van Hoeven N, Glaser L, Solorzano A, Pappas C, Cox NJ, Swayne DE, Palese P, Katz JM, Garcia-Sastre A. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 2007;315:655–659. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- 62.Gamblin SJ, Haire LF, Russell RJ, Stevens DJ, Xiao B, Ha Y, Vasisht N, Steinhauer DA, Daniels RS, Elliot A, Wiley DC, Skehel JJ. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science. 2004;303:1838–1842. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- 63.Van Hoeven N, Pappas C, Belser JA, Maines TR, Zeng H, Garcia-Sastre A, Sasisekharan R, Katz JM, Tumpey TM. Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proc Natl Acad Sci U S A. 2009;106:3366–3371. doi: 10.1073/pnas.0813172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watanabe T, Zhong G, Russell CA, Nakajima N, Hatta M, Hanson A, McBride R, Burke DF, Takahashi K, Fukuyama S, Tomita Y, Maher EA, Watanabe S, Imai M, Neumann G, Hasegawa H, Paulson JC, Smith DJ, Kawaoka Y. Circulating avian influenza viruses closely related to the 1918 virus have pandemic potential. Cell Host Microbe. 2014;15:692–705. doi: 10.1016/j.chom.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, Gustin KM, Pearce MB, Viswanathan K, Shriver ZH, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science. 2009;325:484–487. doi: 10.1126/science.1177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang H, Carney P, Stevens J. Structure and Receptor binding properties of a pandemic H1N1 virus hemagglutinin. PLoS Curr. 2010;2:RRN1152. doi: 10.1371/currents.RRN1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE, Jr., Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Childs RA, Palma AS, Wharton S, Matrosovich T, Liu Y, Chai W, Campanero-Rhodes MA, Zhang Y, Eickmann M, Kiso M, Hay A, Matrosovich M, Feizi T. Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray. Nat Biotechnol. 2009;27:797–799. doi: 10.1038/nbt0909-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang W, Qi J, Shi Y, Li Q, Gao F, Sun Y, Lu X, Lu Q, Vavricka CJ, Liu D, Yan J, Gao GF. Crystal structure of the swine-origin A (H1N1)-2009 influenza A virus hemagglutinin (HA) reveals similar antigenicity to that of the 1918 pandemic virus. Protein Cell. 2010;1:459–467. doi: 10.1007/s13238-010-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Itoh Y, Shinya K, Kiso M, Watanabe T, Sakoda Y, Hatta M, Muramoto Y, Tamura D, Sakai-Tagawa Y, Noda T, Sakabe S, Imai M, Hatta Y, Watanabe S, Li C, Yamada S, Fujii K, Murakami S, Imai H, Kakugawa S, Ito M, Takano R, Iwatsuki-Horimoto K, Shimojima M, Horimoto T, Goto H, Takahashi K, Makino A, Ishigaki H, Nakayama M, Okamatsu M, Warshauer D, Shult PA, Saito R, Suzuki H, Furuta Y, Yamashita M, Mitamura K, Nakano K, Nakamura M, Brockman-Schneider R, Mitamura H, Yamazaki M, Sugaya N, Suresh M, Ozawa M, Neumann G, Gern J, Kida H, Ogasawara K, Kawaoka Y. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460:1021–1025. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K, Garcia-Sastre A, Palese P. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio. 2010;1 doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Steel J, Palese P, Lowen AC. Transmission of a 2009 Pandemic Influenza Virus Shows a Sensitivity to Temperature and Humidity Similar to That of an H3N2 Seasonal Strain. J Virol. 2011;85:1400–1402. doi: 10.1128/JVI.02186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Steel J, Staeheli P, Mubareka S, Garcia-Sastre A, Palese P, Lowen AC. Transmission of pandemic H1N1 influenza virus and impact of prior exposure to seasonal strains or interferon treatment. J Virol. 2010;84:21–26. doi: 10.1128/JVI.01732-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kilander A, Rykkvin R, Dudman SG, Hungnes O. Observed association between the HA1 mutation D222G in the 2009 pandemic influenza A(H1N1) virus and severe clinical outcome, Norway 2009-2010. Euro Surveill. 2010;15 doi: 10.2807/ese.15.09.19498-en. [DOI] [PubMed] [Google Scholar]
- 75.Mak GC, Au KW, Tai LS, Chuang KC, Cheng KC, Shiu TC, Lim W. Association of D222G substitution in haemagglutinin of 2009 pandemic influenza A (H1N1) with severe disease. Euro Surveill. 2010;15 [PubMed] [Google Scholar]
- 76.Glinsky GV. Genomic analysis of pandemic (H1N1) 2009 reveals association of increasing disease severity with emergence of novel hemagglutinin mutations. Cell Cycle. 2010;9:958–970. doi: 10.4161/cc.9.5.10913. [DOI] [PubMed] [Google Scholar]
- 77.Potdar VA, Chadha MS, Jadhav SM, Mullick J, Cherian SS, Mishra AC. Genetic characterization of the influenza A pandemic (H1N1) 2009 virus isolates from India. PLoS One. 2010;5:e9693. doi: 10.1371/journal.pone.0009693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miller RR, MacLean AR, Gunson RN, Carman WF. Occurrence of haemagglutinin mutation D222G in pandemic influenza A(H1N1) infected patients in the West of Scotland, United Kingdom, 2009-10. Euro Surveill. 2010;15 [PubMed] [Google Scholar]
- 79.Anton A, Marcos MA, Martinez MJ, Ramon S, Martinez A, Cardenosa N, Godoy P, Torner N, De Molina P, Isanta R, Jimenez de Anta MT, Pumarola T. D225G mutation in the hemagglutinin protein found in 3 severe cases of 2009 pandemic influenza A (H1N1) in Spain. Diagn Microbiol Infect Dis. 2010;67:207–208. doi: 10.1016/j.diagmicrobio.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 80.Chen H, Wen X, To KK, Wang P, Tse H, Chan JF, Tsoi HW, Fung KS, Tse CW, Lee RA, Chan KH, Yuen KY. Quasispecies of the D225G substitution in the hemagglutinin of pandemic influenza A(H1N1) 2009 virus from patients with severe disease in Hong Kong, China. J Infect Dis. 2010;201:1517–1521. doi: 10.1086/652661. [DOI] [PubMed] [Google Scholar]
- 81.Preliminary review of D222G amino acid substitution in the haemagglutinin of pandemic influenza A (H1N1) 2009 viruses. Wkly Epidemiol Rec. 2010;85:21–22. [PubMed] [Google Scholar]
- 82.Chutinimitkul S, Herfst S, Steel J, Lowen AC, Ye J, van Riel D, Schrauwen EJ, Bestebroer TM, Koel B, Burke DF, Sutherland-Cash KH, Whittleston CS, Russell CA, Wales DJ, Smith DJ, Jonges M, Meijer A, Koopmans M, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Garcia-Sastre A, Perez DR, Fouchier RA. Virulence-associated substitution D222G in the hemagglutinin of 2009 pandemic influenza A(H1N1) virus affects receptor binding. J Virol. 2010;84:11802–11813. doi: 10.1128/JVI.01136-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 84.Naeve CW, Hinshaw VS, Webster RG. Mutations in the hemagglutinin receptor-binding site can change the biological properties of an influenza virus. J Virol. 1984;51:567–569. doi: 10.1128/jvi.51.2.567-569.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Belser JA, Blixt O, Chen LM, Pappas C, Maines TR, Van Hoeven N, Donis R, Busch J, McBride R, Paulson JC, Katz JM, Tumpey TM. Contemporary North American influenza H7 viruses possess human receptor specificity: Implications for virus transmissibility. Proc Natl Acad Sci U S A. 2008;105:7558–7563. doi: 10.1073/pnas.0801259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Belser JA, Gustin KM, Maines TR, Pantin-Jackwood MJ, Katz JM, Tumpey TM. Influenza virus respiratory infection and transmission following ocular inoculation in ferrets. PLoS Pathog. 2012;8:e1002569. doi: 10.1371/journal.ppat.1002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Belser JA, Davis CT, Balish A, Edwards LE, Zeng H, Maines TR, Gustin KM, Martinez IL, Fasce R, Cox NJ, Katz JM, Tumpey TM. Pathogenesis, transmissibility, and ocular tropism of a highly pathogenic avian influenza A (H7N3) virus associated with human conjunctivitis. J Virol. 2013;87:5746–5754. doi: 10.1128/JVI.00154-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gambaryan AS, Matrosovich TY, Philipp J, Munster VJ, Fouchier RA, Cattoli G, Capua I, Krauss SL, Webster RG, Banks J, Bovin NV, Klenk HD, Matrosovich MN. Receptor-binding profiles of H7 subtype influenza viruses in different host species. J Virol. 2012;86:4370–4379. doi: 10.1128/JVI.06959-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang H, Carney PJ, Chang JC, Villanueva JM, Stevens J. Structural analysis of the hemagglutinin from the recent 2013 H7N9 influenza virus. J Virol. 2013;87:12433–12446. doi: 10.1128/JVI.01854-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Srinivasan K, Raman R, Jayaraman A, Viswanathan K, Sasisekharan R. Quantitative description of glycan-receptor binding of influenza A virus H7 hemagglutinin. PLoS One. 2013;8:e49597. doi: 10.1371/journal.pone.0049597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nidom CA, Takano R, Yamada S, Sakai-Tagawa Y, Daulay S, Aswadi D, Suzuki T, Suzuki Y, Shinya K, Iwatsuki-Horimoto K, Muramoto Y, Kawaoka Y. Influenza A (H5N1) viruses from pigs, Indonesia. Emerg Infect Dis. 2010;16:1515–1523. doi: 10.3201/eid1610.100508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ramos I, Krammer F, Hai R, Aguilera D, Bernal-Rubio D, Steel J, Garcia-Sastre A, Fernandez-Sesma A. H7N9 influenza viruses interact preferentially with alpha2,3-linked sialic acids and bind weakly to alpha2,6-linked sialic acids. J Gen Virol. 2013;94:2417–2423. doi: 10.1099/vir.0.056184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van Riel D, Leijten LM, de Graaf M, Siegers JY, Short KR, Spronken MI, Schrauwen EJ, Fouchier RA, Osterhaus AD, Kuiken T. Novel avian-origin influenza A (H7N9) virus attaches to epithelium in both upper and lower respiratory tract of humans. Am J Pathol. 2013;183:1137–1143. doi: 10.1016/j.ajpath.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shi Y, Zhang W, Wang F, Qi J, Wu Y, Song H, Gao F, Bi Y, Zhang Y, Fan Z, Qin C, Sun H, Liu J, Haywood J, Liu W, Gong W, Wang D, Shu Y, Wang Y, Yan J, Gao GF. Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science. 2013;342:243–247. doi: 10.1126/science.1242917. [DOI] [PubMed] [Google Scholar]
- 95.Watanabe T, Kiso M, Fukuyama S, Nakajima N, Imai M, Yamada S, Murakami S, Yamayoshi S, Iwatsuki-Horimoto K, Sakoda Y, Takashita E, McBride R, Noda T, Hatta M, Imai H, Zhao D, Kishida N, Shirakura M, de Wies RP, Shichinohe S, Okamatsu M, Tamura T, Tomita Y, Fujimoto N, Goto K, Katsura H, Kawakami E, Ishikawa I, Watanabe S, Ito M, Sakai-Tagawa Y, Sugita Y, Uraki R, Yamaji R, Eisfeld AJ, Zhong G, Fan S, Ping J, Maher EA, Hanson A, Uchida Y, Saito T, Ozawa M, Neumann G, Kida H, Odagiri T, Paulson JC, Hasegawa H, Tashiro M, Kawaoka Y. Characterization of H7N9 influenza A viruses isolated from humans. Nature. 2013;501:551–555. doi: 10.1038/nature12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xiong X, Coombs PJ, Martin SR, Liu J, Xiao H, McCauley JW, Locher K, Walker PA, Collins PJ, Kawaoka Y, Skehel JJ, Gamblin SJ. Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature. 2013;497:392–396. doi: 10.1038/nature12144. [DOI] [PubMed] [Google Scholar]
- 97.Zhou J, Wang D, Gao R, Zhao B, Song J, Qi X, Zhang Y, Shi Y, Yang L, Zhu W, Bai T, Qin K, Lan Y, Zou S, Guo J, Dong J, Dong L, Zhang Y, Wei H, Li X, Lu J, Liu L, Zhao X, Li X, Huang W, Wen L, Bo H, Xin L, Chen Y, Xu C, Pei Y, Yang Y, Zhang X, Wang S, Feng Z, Han J, Yang W, Gao GF, Wu G, Li D, Wang Y, Shu Y. Biological features of novel avian influenza A (H7N9) virus. Nature. 2013;499:500–503. doi: 10.1038/nature12379. [DOI] [PubMed] [Google Scholar]
- 98.Tharakaraman K, Jayaraman A, Raman R, Viswanathan K, Stebbins NW, Johnson D, Shriver Z, Sasisekharan V, Sasisekharan R. Glycan receptor binding of the influenza A virus H7N9 hemagglutinin. Cell. 2013;153:1486–1493. doi: 10.1016/j.cell.2013.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu L, Bao L, Deng W, Dong L, Zhu H, Chen T, Lv Q, Li F, Yuan J, Xiang Z, Gao K, Xu Y, Huang L, Li Y, Liu J, Yao Y, Yu P, Li X, Huang W, Zhao X, Lan Y, Guo J, Yong W, Wei Q, Chen H, Zhang L, Qin C. Novel avian-origin human influenza A(H7N9) can be transmitted between ferrets via respiratory droplets. J Infect Dis. 2014;209:551–556. doi: 10.1093/infdis/jit474. [DOI] [PubMed] [Google Scholar]
- 100.Yen HL, Zhou J, Choy KT, Sia SF, Teng O, Ng IH, Fang VJ, Hu Y, Wang W, Cowling BJ, Nicholls JM, Guan Y, Peiris JS. The R292K Mutation That Confers Resistance to Neuraminidase Inhibitors Leads to Competitive Fitness Loss of A/Shanghai/1/2013 (H7N9) Influenza Virus in Ferrets. J Infect Dis. 2014 doi: 10.1093/infdis/jiu353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Richard M, Schrauwen EJ, de Graaf M, Bestebroer TM, Spronken MI, van Boheemen S, de Meulder D, Lexmond P, Linster M, Herfst S, Smith DJ, van den Brand JM, Burke DF, Kuiken T, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Limited airborne transmission of H7N9 influenza A virus between ferrets. Nature. 2013;501:560–563. doi: 10.1038/nature12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang Q, Shi J, Deng G, Guo J, Zeng X, He X, Kong H, Gu C, Li X, Liu J, Wang G, Chen Y, Liu L, Liang L, Li Y, Fan J, Wang J, Li W, Guan L, Li Q, Yang H, Chen P, Jiang L, Guan Y, Xin X, Jiang Y, Tian G, Wang X, Qiao C, Li C, Bu Z, Chen H. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science. 2013;341:410–414. doi: 10.1126/science.1240532. [DOI] [PubMed] [Google Scholar]
- 103.Belser JA, Gustin KM, Pearce MB, Maines TR, Zeng H, Pappas C, Sun X, Carney PJ, Villanueva JM, Stevens J, Katz JM, Tumpey TM. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature. 2013;501:556–559. doi: 10.1038/nature12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhu H, Wang D, Kelvin DJ, Li L, Zheng Z, Yoon SW, Wong SS, Farooqui A, Wang J, Banner D, Chen R, Zheng R, Zhou J, Zhang Y, Hong W, Dong W, Cai Q, Roehrl MH, Huang SS, Kelvin AA, Yao T, Zhou B, Chen X, Leung GM, Poon LL, Webster RG, Webby RJ, Peiris JS, Guan Y, Shu Y. Infectivity, Transmission, and Pathology of Human H7N9 Influenza in Ferrets and Pigs. Science. 2013 doi: 10.1126/science.1239844. [DOI] [PubMed] [Google Scholar]
- 105.Gabbard JD, Dlugolenski D, Van Riel D, Marshall N, Galloway SE, Howerth EW, Campbell PJ, Jones C, Johnson S, Byrd-Leotis L, Steinhauer DA, Kuiken T, Tompkins SM, Tripp R, Lowen AC, Steel J. Novel H7N9 influenza virus shows low infectious dose, high growth rate, and efficient contact transmission in the guinea pig model. J Virol. 2014;88:1502–1512. doi: 10.1128/JVI.02959-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hai R, Schmolke M, Leyva-Grado VH, Thangavel RR, Margine I, Jaffe EL, Krammer F, Solorzano A, Garcia-Sastre A, Palese P, Bouvier NM. Influenza A(H7N9) virus gains neuraminidase inhibitor resistance without loss of in vivo virulence or transmissibility. Nat Commun. 2013;4:2854. doi: 10.1038/ncomms3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu T, Bi Z, Wang X, Li Z, Ding S, Bi Z, Wang L, Pei Y, Song S, Zhang S, Wang J, Sun D, Pang B, Sun L, Jiang X, Lei J, Yuan Q, Kou Z, Yang B, Shu Y, Yang L, Li X, Lu K, Liu J, Zhang T, Xu A. One family cluster of avian influenza A(H7N9) virus infection in Shandong, China. BMC Infect Dis. 2014;14:98. doi: 10.1186/1471-2334-14-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Qi X, Qian YH, Bao CJ, Guo XL, Cui LB, Tang FY, Ji H, Huang Y, Cai PQ, Lu B, Xu K, Shi C, Zhu FC, Zhou MH, Wang H. Probable person to person transmission of novel avian influenza A (H7N9) virus in Eastern China, 2013: epidemiological investigation. BMJ. 2013;347:f4752. doi: 10.1136/bmj.f4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sutton TC, Finch C, Shao H, Angel M, Chen H, Capua I, Cattoli G, Monne I, Perez DR. Airborne transmission of highly pathogenic H7N1 influenza virus in ferrets. J Virol. 2014;88:6623–6635. doi: 10.1128/JVI.02765-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Matrosovich M, Zhou N, Kawaoka Y, Webster R. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J Virol. 1999;73:1146–1155. doi: 10.1128/jvi.73.2.1146-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Watanabe Y, Ibrahim MS, Suzuki Y, Ikuta K. The changing nature of avian influenza A virus (H5N1). Trends Microbiol. 2012;20:11–20. doi: 10.1016/j.tim.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 112.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 113.Auewarakul P, Suptawiwat O, Kongchanagul A, Sangma C, Suzuki Y, Ungchusak K, Louisirirotchanakul S, Lerdsamran H, Pooruk P, Thitithanyanont A, Pittayawonganon C, Guo CT, Hiramatsu H, Jampangern W, Chunsutthiwat S, Puthavathana P. An avian influenza H5N1 virus that binds to a human-type receptor. J Virol. 2007;81:9950–9955. doi: 10.1128/JVI.00468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kongchanagul A, Suptawiwat O, Kanrai P, Uiprasertkul M, Puthavathana P, Auewarakul P. Positive selection at the receptor-binding site of haemagglutinin H5 in viral sequences derived from human tissues. J Gen Virol. 2008;89:1805–1810. doi: 10.1099/vir.0.2008/002469-0. [DOI] [PubMed] [Google Scholar]
- 115.Yen HL, Lipatov AS, Ilyushina NA, Govorkova EA, Franks J, Yilmaz N, Douglas A, Hay A, Krauss S, Rehg JE, Hoffmann E, Webster RG. Inefficient transmission of H5N1 influenza viruses in a ferret contact model. J Virol. 2007;81:6890–6898. doi: 10.1128/JVI.00170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Watanabe Y, Ibrahim MS, Ellakany HF, Kawashita N, Mizuike R, Hiramatsu H, Sriwilaijaroen N, Takagi T, Suzuki Y, Ikuta K. Acquisition of human-type receptor binding specificity by new H5N1 influenza virus sublineages during their emergence in birds in Egypt. PLoS Pathog. 2011;7:e1002068. doi: 10.1371/journal.ppat.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chutinimitkul S, van Riel D, Munster VJ, van den Brand JM, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA, de Wit E. In vitro assessment of attachment pattern and replication efficiency of H5N1 influenza A viruses with altered receptor specificity. J Virol. 2010;84:6825–6833. doi: 10.1128/JVI.02737-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Maines TR, Chen LM, Van Hoeven N, Tumpey TM, Blixt O, Belser JA, Gustin KM, Pearce MB, Pappas C, Stevens J, Cox NJ, Paulson JC, Raman R, Sasisekharan R, Katz JM, Donis RO. Effect of receptor binding domain mutations on receptor binding and transmissibility of avian influenza H5N1 viruses. Virology. 2011;413:139–147. doi: 10.1016/j.virol.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stevens J, Blixt O, Chen LM, Donis RO, Paulson JC, Wilson IA. Recent avian H5N1 viruses exhibit increased propensity for acquiring human receptor specificity. J Mol Biol. 2008;381:1382–1394. doi: 10.1016/j.jmb.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ayora-Talavera G, Shelton H, Scull MA, Ren J, Jones IM, Pickles RJ, Barclay WS. Mutations in H5N1 influenza virus hemagglutinin that confer binding to human tracheal airway epithelium. PLoS One. 2009;4:e7836. doi: 10.1371/journal.pone.0007836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lowen AC, Mubareka S, Tumpey TM, Garcia-Sastre A, Palese P. The guinea pig as a transmission model for human influenza viruses. Proc Natl Acad Sci U S A. 2006;103:9988–9992. doi: 10.1073/pnas.0604157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Maines TR, Chen LM, Matsuoka Y, Chen H, Rowe T, Ortin J, Falcon A, Nguyen TH, Mai le Q, Sedyaningsih ER, Harun S, Tumpey TM, Donis RO, Cox NJ, Subbarao K, Katz JM. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci U S A. 2006;103:12121–12126. doi: 10.1073/pnas.0605134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen LM, Blixt O, Stevens J, Lipatov AS, Davis CT, Collins BE, Cox NJ, Paulson JC, Donis RO. In vitro evolution of H5N1 avian influenza virus toward human-type receptor specificity. Virology. 2012;422:105–113. doi: 10.1016/j.virol.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen LM, Davis CT, Zhou H, Cox NJ, Donis RO. Genetic compatibility and virulence of reassortants derived from contemporary avian H5N1 and human H3N2 influenza A viruses. PLoS Pathog. 2008;4:e1000072. doi: 10.1371/journal.ppat.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li C, Hatta M, Nidom CA, Muramoto Y, Watanabe S, Neumann G, Kawaoka Y. Reassortment between avian H5N1 and human H3N2 influenza viruses creates hybrid viruses with substantial virulence. Proc Natl Acad Sci U S A. 2010;107:4687–4692. doi: 10.1073/pnas.0912807107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang Y, Zhang Q, Kong H, Jiang Y, Gao Y, Deng G, Shi J, Tian G, Liu L, Liu J, Guan Y, Bu Z, Chen H. H5N1 Hybrid Viruses Bearing 2009/H1N1 Virus Genes Transmit in Guinea Pigs by Respiratory Droplet. Science. 2013 doi: 10.1126/science.1229455. [DOI] [PubMed] [Google Scholar]
- 129.Jackson S, Van Hoeven N, Chen LM, Maines TR, Cox NJ, Katz JM, Donis RO. Reassortment between avian H5N1 and human H3N2 influenza viruses in ferrets: a public health risk assessment. J Virol. 2009;83:8131–8140. doi: 10.1128/JVI.00534-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schrauwen EJ, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA, Herfst S. Reassortment between Avian H5N1 and human influenza viruses is mainly restricted to the matrix and neuraminidase gene segments. PLoS One. 2013;8:e59889. doi: 10.1371/journal.pone.0059889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cline TD, Karlsson EA, Freiden P, Seufzer BJ, Rehg JE, Webby RJ, Schultz-Cherry S. Increased pathogenicity of a reassortant 2009 pandemic H1N1 influenza virus containing an H5N1 hemagglutinin. Journal of virology. 2011;85:12262–12270. doi: 10.1128/JVI.05582-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Linster M, van Boheemen S, de Graaf M, Schrauwen EJ, Lexmond P, Manz B, Bestebroer TM, Baumann J, van Riel D, Rimmelzwaan GF, Osterhaus AD, Matrosovich M, Fouchier RA, Herfst S. Identification, characterization, and natural selection of mutations driving airborne transmission of A/H5N1 virus. Cell. 2014;157:329–339. doi: 10.1016/j.cell.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lu X, Shi Y, Zhang W, Zhang Y, Qi J, Gao GF. Structure and receptor-binding properties of an airborne transmissible avian influenza A virus hemagglutinin H5 (VN1203mut). Protein Cell. 2013;4:502–511. doi: 10.1007/s13238-013-3906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang W, Shi Y, Lu X, Shu Y, Qi J, Gao GF. An Airborne Transmissible Avian Influenza H5 Hemagglutinin Seen at the Atomic Level. Science. 2013 doi: 10.1126/science.1236787. [DOI] [PubMed] [Google Scholar]
- 135.Steel J, Lowen AC, Mubareka S, Palese P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 2009;5:e1000252. doi: 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gao Y, Zhang Y, Shinya K, Deng G, Jiang Y, Li Z, Guan Y, Tian G, Li Y, Shi J, Liu L, Zeng X, Bu Z, Xia X, Kawaoka Y, Chen H. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog. 2009;5:e1000709. doi: 10.1371/journal.ppat.1000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Saito T, Lim W, Suzuki T, Suzuki Y, Kida H, Nishimura SI, Tashiro M. Characterization of a human H9N2 influenza virus isolated in Hong Kong. Vaccine. 2001;20:125–133. doi: 10.1016/s0264-410x(01)00279-1. [DOI] [PubMed] [Google Scholar]
- 138.Butt KM, Smith GJ, Chen H, Zhang LJ, Leung YH, Xu KM, Lim W, Webster RG, Yuen KY, Peiris JS, Guan Y. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J Clin Microbiol. 2005;43:5760–5767. doi: 10.1128/JCM.43.11.5760-5767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li C, Chen H. Enhancement of Influenza Virus Transmission by Gene Reassortment. In: Compans RW, Oldstone MB, editors. Influenza Patlgmesis and Control - Volume I. Springer; Heidelberg, New York, Dordrecht, London: 2014. pp. 185–204. [DOI] [PubMed] [Google Scholar]
- 140.Neumann G, Green MA, Macken CA. Evolution of highly pathogenic avian H5N1 influenza viruses and the emergence of dominant variants. J Gen Virol. 2010;91:1984–1995. doi: 10.1099/vir.0.020750-0. [DOI] [PubMed] [Google Scholar]
- 141.Guan Y, Shortridge KF, Krauss S, Webster RG. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci U S A. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Guan Y, Poon LL, Cheung CY, Ellis TM, Lim W, Lipatov AS, Chan KH, Sturm-Ramirez KM, Cheung CL, Leung YH, Yuen KY, Webster RG, Peiris JS. H5N1 influenza: a protean pandemic threat. Proc Natl Acad Sci U S A. 2004;101:8156–8161. doi: 10.1073/pnas.0402443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Guan Y, Peiris JS, Lipatov AS, Ellis TM, Dyrting KC, Krauss S, Zhang LJ, Webster RG, Shortridge KF. Emergence of multiple genotypes of H5N1 avian influenza viruses in Hong Kong SAR. Proc Natl Acad Sci U S A. 2002;99:8950–8955. doi: 10.1073/pnas.132268999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li KS, Guan Y, Wang J, Smith GJ, Xu KM, Duan L, Rahardjo AP, Puthavathana P, Buranathai C, Nguyen TD, Estoepangestie AT, Chaisingh A, Auewarakul P, Long HT, Hanh NT, Webby RJ, Poon LL, Chen H, Shortridge KF, Yuen KY, Webster RG, Peiris JS. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- 145.Duan L, Bahl J, Smith GJ, Wang J, Vijaykrishna D, Zhang LJ, Zhang JX, Li KS, Fan XH, Cheung CL, Huang K, Poon LL, Shortridge KF, Webster RG, Peiris JS, Chen H, Guan Y. The development and genetic diversity of H5N1 influenza virus in China, 1996-2006. Virology. 2008;380:243–254. doi: 10.1016/j.virol.2008.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Freidl GS, Meijer A, de Bruin E, de Nardi M, Munoz O, Capua I, Breed AC, Harris K, Hill A, Kosmider R, Banks J, von Dobschuetz S, Stark K, Wieland B, Stevens K, van der Werf S, Enouf V, van der Meulen K, Van Reeth K, Dauphin G, Koopmans M, Consortium F. Influenza at the animal-human interface: a review of the literature for virological evidence of human infection with swine or avian influenza viruses other than A(H5N1). Euro Surveill. 2014;19 doi: 10.2807/1560-7917.es2014.19.18.20793. [DOI] [PubMed] [Google Scholar]
- 147.Matrosovich MN, Krauss S, Webster RG. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology. 2001;281:156–162. doi: 10.1006/viro.2000.0799. [DOI] [PubMed] [Google Scholar]
- 148.Wan H, Perez DR. Amino acid 226 in the hemagglutinin of H9N2 influenza viruses determines cell tropism and replication in human airway epithelial cells. J Virol. 2007;81:5181–5191. doi: 10.1128/JVI.02827-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wan H, Sorrell EM, Song H, Hossain MJ, Ramirez-Nieto G, Monne I, Stevens J, Cattoli G, Capua I, Chen LM, Donis RO, Busch J, Paulson JC, Brockwell C, Webby R, Blanco J, Al-Natour MQ, Perez DR. Replication and transmission of H9N2 influenza viruses in ferrets: evaluation of pandemic potential. PLoS One. 2008;3:e2923. doi: 10.1371/journal.pone.0002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sorrell EM, Wan H, Araya Y, Song H, Perez DR. Minimal molecular constraints for respiratory droplet transmission of an avian-human H9N2 influenza A virus. Proc Natl Acad Sci U S A. 2009;106:7565–7570. doi: 10.1073/pnas.0900877106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kimble JB, Sorrell E, Shao H, Martin PL, Perez DR. Compatibility of H9N2 avian influenza surface genes and 2009 pandemic H1N1 internal genes for transmission in the ferret model. Proc Natl Acad Sci U S A. 2011;108:12084–12088. doi: 10.1073/pnas.1108058108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang G, Deng G, Shi J, Luo W, Zhang G, Zhang Q, Liu L, Jiang Y, Li C, Sriwilaijaroen N, Hiramatsu H, Suzuki Y, Kawaoka Y, Chen H. H6 influenza viruses pose a potential threat to human health. J Virol. 2014;88:3953–3964. doi: 10.1128/JVI.03292-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lamblin G, Roussel P. Airway mucins and their role in defence against micro-organisms. Respiratory medicine. 1993;87:421–426. doi: 10.1016/0954-6111(93)90067-a. [DOI] [PubMed] [Google Scholar]
- 154.Wright PF, Neumann G, Kawaoka Y. Orthomyxoviruses. In: Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B, editors. Fields Virology. Sixth ed One. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2013. pp. 1186–1243. [Google Scholar]
- 155.Wang W, Lu B, Zhou H, Suguitan AL, Jr., Cheng X, Subbarao K, Kemble G, Jin H. Glycosylation at 158N of the hemagglutinin protein and receptor binding specificity synergistically affect the antigenicity and immunogenicity of a live attenuated H5N1 A/Vietnam/1203/2004 vaccine virus in ferrets. J Virol. 2010;84:6570–6577. doi: 10.1128/JVI.00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Neumann G, Macken CA, Karasin AI, Fouchier RA, Kawaoka Y. Egyptian H5N1 influenza viruses-cause for concern? PLoS Pathog. 2012;8:e1002932. doi: 10.1371/journal.ppat.1002932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.de Vries RP, Zhu X, McBride R, Rigter A, Hanson A, Zhong G, Hatta M, Xu R, Yu W, Kawaoka Y, de Haan CA, Wilson IA, Paulson JC. Hemagglutinin Receptor Specificity and Structural Analyses of Respiratory Droplet-Transmissible H5N1 Viruses. J Virol. 2014;88:768–773. doi: 10.1128/JVI.02690-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Russell CJ. Acid-Induced Membrane Fusion by the Hemagglutinin Protein and Its Role in Influenza Virus Biology. In: Compans RW, Oldstone MB, editors. Influenza Pathogenesis and Control - Volume 1. Springer; Heidelberg, New York, Dordrecht, London: 2014. pp. 93–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Reed ML, Yen HL, DuBois RM, Bridges OA, Salomon R, Webster RG, Russell CJ. Amino acid residues in the fusion peptide pocket regulate the pH of activation of the H5N1 influenza virus hemagglutinin protein. J Virol. 2009;83:3568–3580. doi: 10.1128/JVI.02238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Reed ML, Bridges OA, Seiler P, Kim JK, Yen HL, Salomon R, Govorkova EA, Webster RG, Russell CJ. The pH of activation of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity and transmissibility in ducks. J Virol. 2010;84:1527–1535. doi: 10.1128/JVI.02069-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zaraket H, Bridges OA, Russell CJ. The pH of activation of the hemagglutinin protein regulates H5N1 influenza virus replication and pathogenesis in mice. J Virol. 2013;87:4826–4834. doi: 10.1128/JVI.03110-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zaraket H, Bridges OA, Duan S, Baranovich T, Yoon SW, Reed ML, Salomon R, Webby RJ, Webster RG, Russell CJ. Increased acid stability of the hemagglutinin protein enhances H5N1 influenza virus growth in the upper respiratory tract but is insufficient for transmission in ferrets. J Virol. 2013 doi: 10.1128/JVI.01175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.DuBois RM, Zaraket H, Reddivari M, Heath RJ, White SW, Russell CJ. Acid stability of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity. PLoS Pathog. 2011;7:e1002398. doi: 10.1371/journal.ppat.1002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Baum LG, Paulson JC. The N2 neuraminidase of human influenza virus has acquired a substrate specificity complementary to the hemagglutinin receptor specificity. Virology. 1991;180:10–15. doi: 10.1016/0042-6822(91)90003-t. [DOI] [PubMed] [Google Scholar]
- 165.Kobasa D, Kodihalli S, Luo M, Castrucci MR, Donatelli I, Suzuki Y, Suzuki T, Kawaoka Y. Amino acid residues contributing to the substrate specificity of the influenza A virus neuraminidase. J Virol. 1999;73:6743–6751. doi: 10.1128/jvi.73.8.6743-6751.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Garcia JM, Lai JC, Haselhorst T, Choy KT, Yen HL, Peiris JS, von Itzstein M, Nicholls JM. Investigation of the binding and cleavage characteristics of N1 neuraminidases from avian, seasonal, and pandemic influenza viruses using saturation transfer difference nuclear magnetic resonance. Influenza and other respiratory viruses. 2014;8:235–242. doi: 10.1111/irv.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yen HL, Liang CH, Wu CY, Forrest HL, Ferguson A, Choy KT, Jones J, Wong DD, Cheung PP, Hsu CH, Li OT, Yuen KM, Chan RW, Poon LL, Chan MC, Nicholls JM, Krauss S, Wong CH, Guan Y, Webster RG, Webby RJ, Peiris M. Hemagglutinin-neuraminidase balance confers respiratory-droplet transmissibility of the pandemic H1N1 influenza virus in ferrets. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14264–14269. doi: 10.1073/pnas.1111000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhou H, Yu Z, Hu Y, Tu J, Zou W, Peng Y, Zhu J, Li Y, Zhang A, Ye Z, Chen H, Jin M. The special neuraminidase stalk-motif responsible for increased virulence and pathogenesis of H5N1 influenza A virus. PLoS One. 2009;4:e6277. doi: 10.1371/journal.pone.0006277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Munier S, Larcher T, Cormier-Aline F, Soubieux D, Su B, Guigand L, Labrosse B, Cherel Y, Quere P, Marc D, Naffakh N. A genetically engineered waterfowl influenza virus with a deletion in the stalk of the neuraminidase has increased virulence for chickens. J Virol. 2010;84:940–952. doi: 10.1128/JVI.01581-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Banks J, Speidel ES, Moore E, Plowright L, Piccirillo A, Capua I, Cordioli P, Fioretti A, Alexander DJ. Changes in the haemagglutinin and the neuraminidase genes prior to the emergence of highly pathogenic H7N1 avian influenza viruses in Italy. Arch Virol. 2001;146:963–973. doi: 10.1007/s007050170128. [DOI] [PubMed] [Google Scholar]
- 171.Campitelli L, Mogavero E, De Marco MA, Delogu M, Puzelli S, Frezza F, Facchini M, Chiapponi C, Foni E, Cordioli P, Webby R, Barigazzi G, Webster RG, Donatelli I. Interspecies transmission of an H7N3 influenza virus from wild birds to intensively reared domestic poultry in Italy. Virology. 2004;323:24–36. doi: 10.1016/j.virol.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 172.Giannecchini S, Campitelli L, Calzoletti L, De Marco MA, Azzi A, Donatelli I. Comparison of in vitro replication features of H7N3 influenza viruses from wild ducks and turkeys: potential implications for interspecies transmission. J Gen Virol. 2006;87:171–175. doi: 10.1099/vir.0.81187-0. [DOI] [PubMed] [Google Scholar]
- 173.Li J, Zu Dohna H, Cardona CJ, Miller J, Carpenter TE. Emergence and genetic variation of neuraminidase stalk deletions in avian influenza viruses. PLoS One. 2011;6:e14722. doi: 10.1371/journal.pone.0014722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yamada S, Shinya K, Takada A, Ito T, Suzuki T, Suzuki Y, Le QM, Ebina M, Kasai N, Kida H, Horimoto T, Rivailler P, Chen LM, Donis RO, Kawaoka Y. Adaptation of a duck influenza A virus in quail. J Virol. 2012;86:1411–1420. doi: 10.1128/JVI.06100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Blumenkrantz D, Roberts KL, Shelton H, Lycett S, Barclay WS. The short stalk length of highly pathogenic avian influenza H5N1 virus neuraminidase limits transmission of pandemic H1N1 virus in ferrets. J Virol. 2013;87:10539–10551. doi: 10.1128/JVI.00967-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Subbarao EK, Kawaoka Y, Murphy BR. Rescue of an influenza A virus wild-type PB2 gene and a mutant derivative bearing a site-specific temperature-sensitive and attenuating mutation. J Virol. 1993;67:7223–7228. doi: 10.1128/jvi.67.12.7223-7228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 178.Hatta M, Hatta Y, Kim JH, Watanabe S, Shinya K, Nguyen T, Lien PS, Le QM, Kawaoka Y. Growth of H5N1 influenza A viruses in the upper respiratory tracts of mice. PLoS Pathog. 2007;3:1374–1379. doi: 10.1371/journal.ppat.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Chen H, Smith GJ, Zhang SY, Qin K, Wang J, Li KS, Webster RG, Peiris JS, Guan Y. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature. 2005;436:191–192. doi: 10.1038/nature03974. [DOI] [PubMed] [Google Scholar]
- 180.Chen H, Li Y, Li Z, Shi J, Shinya K, Deng G, Qi Q, Tian G, Fan S, Zhao H, Sun Y, Kawaoka Y. Properties and dissemination of H5N1 viruses isolated during an influenza outbreak in migratory waterfowl in western China. J Virol. 2006;80:5976–5983. doi: 10.1128/JVI.00110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liu J, Xiao H, Lei F, Zhu Q, Qin K, Zhang XW, Zhang XL, Zhao D, Wang G, Feng Y, Ma J, Liu W, Wang J, Gao GF. Highly pathogenic H5N1 influenza virus infection in migratory birds. Science. 2005;309:1206. doi: 10.1126/science.1115273. [DOI] [PubMed] [Google Scholar]
- 182.Mehle A, Doudna JA. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc Natl Acad Sci U S A. 2009;106:21312–21316. doi: 10.1073/pnas.0911915106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yamada S, Hatta M, Staker BL, Watanabe S, Imai M, Shinya K, Sakai-Tagawa Y, Ito M, Ozawa M, Watanabe T, Sakabe S, Li C, Kim JH, Myler PJ, Phan I, Raymond A, Smith E, Stacy R, Nidom CA, Lank SM, Wiseman RW, Bimber BN, O'Connor DH, Neumann G, Stewart LJ, Kawaoka Y. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS pathogens. 2010;6:e1001034. doi: 10.1371/journal.ppat.1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Li Z, Chen H, Jiao P, Deng G, Tian G, Li Y, Hoffmann E, Webster RG, Matsuoka Y, Yu K. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J Virol. 2005;79:12058–12064. doi: 10.1128/JVI.79.18.12058-12064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gabriel G, Dauber B, Wolff T, Planz O, Klenk HD, Stech J. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc Natl Acad Sci U S A. 2005;102:18590–18595. doi: 10.1073/pnas.0507415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gabriel G, Herwig A, Klenk HD. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog. 2008;4:e11. doi: 10.1371/journal.ppat.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bussey KA, Bousse TL, Desmet EA, Kim B, Takimoto T. PB2 residue 271 plays a key role in enhanced polymerase activity of influenza A viruses in mammalian host cells. J Virol. 2010;84:4395–4406. doi: 10.1128/JVI.02642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang Y, Zhang Q, Gao Y, He X, Kong H, Jiang Y, Guan Y, Xia X, Shu Y, Kawaoka Y, Bu Z, Chen H. Key molecular factors in hemagglutinin and PB2 contribute to efficient transmission of the 2009 H1N1 pandemic influenza virus. J Virol. 2012;86:9666–9674. doi: 10.1128/JVI.00958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pflug A, Guilligay D, Reich S, Cusack S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature. 2014;516:355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 190.Reich S, Guilligay D, Pflug A, Malet H, Berger I, Crepin T, Hart D, Lunardi T, Nanao M, Ruigrok RW, Cusack S. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature. 2014;516:361–366. doi: 10.1038/nature14009. [DOI] [PubMed] [Google Scholar]
- 191.Liu Y, Qin K, Meng G, Zhang J, Zhou J, Zhao G, Luo M, Zheng X. Structural and functional characterization of K339T substitution identified in the PB2 subunit cap-binding pocket of influenza A virus. J Biol Chem. 2013;288:11013–11023. doi: 10.1074/jbc.M112.392878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Fan S, Hatta M, Kim JH, Halfmann P, Imai M, Macken CA, Le MQ, Nguyen T, Neumann G, Kawaoka Y. Novel residues in avian influenza virus PB2 protein affect virulence in mammalian hosts. Nat Commun. 2014;5:5021. doi: 10.1038/ncomms6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yamaji R, Yamada S, Le MQ, Li C, Chen H, Qurnianingsih E, Nidom CA, Ito M, Sakai-Tagawa Y, Kawaoka Y. Identification of PB2 mutations responsible for the efficient replication of H5N1 influenza viruses in human lung epithelial cells. J Virol. 2015 doi: 10.1128/JVI.03328-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mok CK, Yen HL, Yu MY, Yuen KM, Sia SF, Chan MC, Qin G, Tu WW, Peiris JS. Amino acid residues 253 and 591 of the PB2 protein of avian influenza virus A H9N2 contribute to mammalian pathogenesis. Journal of virology. 2011;85:9641–9645. doi: 10.1128/JVI.00702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Manzoor R, Sakoda Y, Nomura N, Tsuda Y, Ozaki H, Okamatsu M, Kida H. PB2 protein of a highly pathogenic avian influenza virus strain A/chicken/Yamaguchi/7/2004 (H5N1) determines its replication potential in pigs. J Virol. 2009;83:1572–1578. doi: 10.1128/JVI.01879-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Leung BW, Chen H, Brownlee GG. Correlation between polymerase activity and pathogenicity in two duck H5N1 influenza viruses suggests that the polymerase contributes to pathogenicity. Virology. 2010;401:96–106. doi: 10.1016/j.virol.2010.01.036. [DOI] [PubMed] [Google Scholar]
- 197.Belser JA, Gustin KM, Maines TR, Blau DM, Zaki SR, Katz JM, Tumpey TM. Pathogenesis and transmission of triple-reassortant swine H1N1 influenza viruses isolated before the 2009 H1N1 pandemic. J Virol. 2011;85:1563–1572. doi: 10.1128/JVI.02231-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Barman S, Krylov PS, Fabrizio TP, Franks J, Turner JC, Seiler P, Wang D, Rehg JE, Erickson GA, Gramer M, Webster RG, Webby RJ. Pathogenicity and transmissibility of North American triple reassortant swine influenza A viruses in ferrets. PLoS Pathog. 2012;8:e1002791. doi: 10.1371/journal.ppat.1002791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pascua PN, Song MS, Lee JH, Baek YH, Kwon HI, Park SJ, Choi EH, Lim GJ, Lee OJ, Kim SW, Kim CJ, Sung MH, Kim MH, Yoon SW, Govorkova EA, Webby RJ, Webster RG, Choi YK. Virulence and transmissibility of H1N2 influenza virus in ferrets imply the continuing threat of triple-reassortant swine viruses. Proc Natl Acad Sci U S A. 2012;109:15900–15905. doi: 10.1073/pnas.1205576109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lakdawala SS, Lamirande EW, Suguitan AL, Jr., Wang W, Santos CP, Vogel L, Matsuoka Y, Lindsley WG, Jin H, Subbarao K. Eurasian-origin gene segments contribute to the transmissibility, aerosol release, and morphology of the 2009 pandemic H1N1 influenza virus. PLoS Pathog. 2011;7:e1002443. doi: 10.1371/journal.ppat.1002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Campbell PJ, Danzy S, Kyriakis CS, Deymier MJ, Lowen AC, Steel J. The M segment of the 2009 pandemic influenza virus confers increased neuraminidase activity, filamentous morphology, and efficient contact transmissibility to A/Puerto Rico/8/1934-based reassortant viruses. J Virol. 2014;88:3802–3814. doi: 10.1128/JVI.03607-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Chou YY, Albrecht RA, Pica N, Lowen AC, Richt JA, Garcia-Sastre A, Palese P, Hai R. The M segment of the 2009 new pandemic H1N1 influenza virus is critical for its high transmission efficiency in the guinea pig model. J Virol. 2011;85:11235–11241. doi: 10.1128/JVI.05794-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ma W, Liu Q, Bawa B, Qiao C, Qi W, Shen H, Chen Y, Ma J, Li X, Webby RJ, Garcia-Sastre A, Richt JA. The neuraminidase and matrix genes of the 2009 pandemic influenza H1N1 virus cooperate functionally to facilitate efficient replication and transmissibility in pigs. The Journal of general virology. 2012;93:1261–1268. doi: 10.1099/vir.0.040535-0. [DOI] [PMC free article] [PubMed] [Google Scholar]