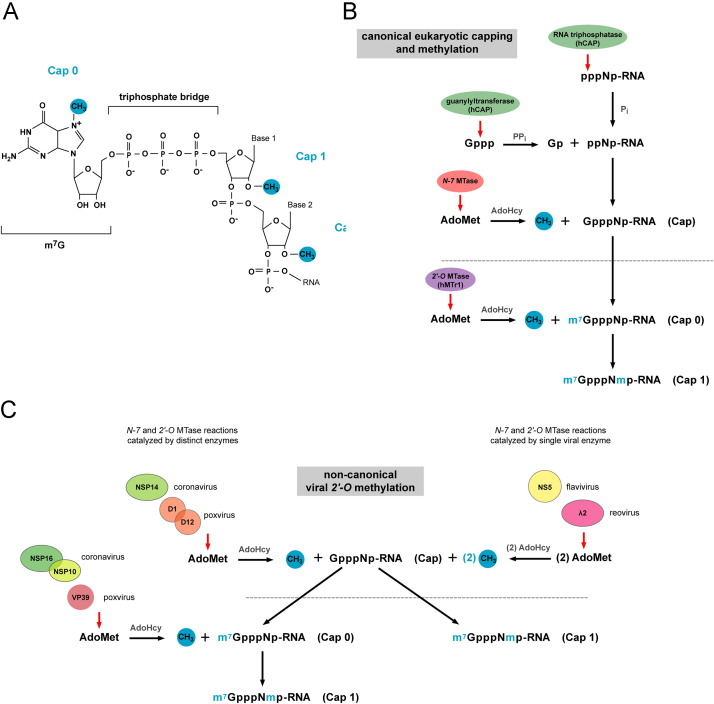
Fig. 1.
Overview of cap 1 structure formation in eukaryotic and viral systems. (A) Chemical structure of the G-cap and methylation sites which form cap 0, cap 1, and cap 2 structures. (B) The eukaryotic canonical capping pathway. Newly transcribed RNA (pppNp-RNA) is cleaved by RNA triphosphatase at the γ-phosphate to yield diphosphate RNA (ppNp-RNA). Guanylyltransferase, which forms part of the bifunctional capping enzyme (hCAP) that also encodes triphosphatase activity, then hydrolyzes the α-phosphate from GTP and transfers the GMP moiety to the ppNp-RNA acceptor to generate GpppNp. GpppNp-RNA is methylated by the N-7 MTase, which transfers a methyl group from an S-adenosyl-l-methionine (AdoMet) donor to the unmethylated G-cap acceptor, producing cap 0 RNA (m7GpppNp) and S-adenosyl-l-homocysteine (AdoHcy) as a by-product. The 2′-O-ribose MTase (hMTr1) then transfers a methyl group from the AdoMet donor to the first and second nucleotides of cap 0 RNA to generate cap 1 and cap 2 structures, respectively. (C) Generation of viral cap 1 structures through non-canonical capping and methylation pathways. Analogous to eukaryotic RNA methylation, some viruses (e.g. coronavirus and poxvirus) catalyze cap 0 and cap 1 formation separately via two distinct enzymes or enzymes complexes (left). In contrast, other viruses (e.g. flavivirus and reovirus) catalyze N-7 and 2′-O methylation sequentially via a single enzyme (right). Although the cap 1 end product is identical between host and viral transcripts structural differences in the viral and eukaryotic enzymes that catalyze these reactions and their distinct modes of ligand binding make viral 2′-O MTases potential targets for inhibitor design.