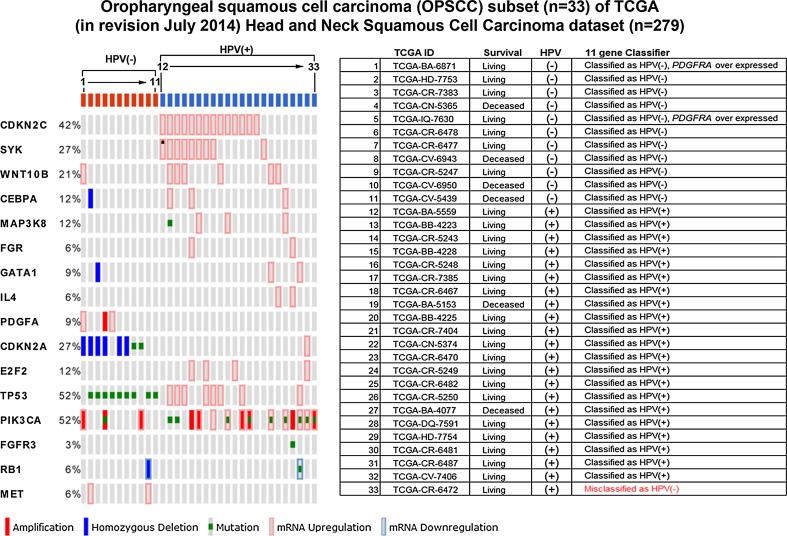
Fig. 5.
Eleven gene HPV classifier correctly identifies 97 % (32/33) OPSCC samples in the TCGA HNSCC dataset. OncoPrint output of the TCGA OPSCC subset using our 11 gene expression HPV classifier and clinically relevant mutated gene list (PIK3CA, TP53, FGFR3, RB1 and MET). Over expressed genes used to predict HPV status in our training set shown in Table 3 were parsed to 11 genes and applied to the 33 sample OPSCC subset of the full 279 specimen TCGA HNSCC dataset using cBioPortal. The 11 gene classifier required overexpression of one or more of ten genes (CDKN2C, SYK, WNT10B, CEBPA, MAP3K8, FGR, GATA1, IL4, CDKN2A and E2F2) for HPV-positive status and PDGFA overexpression or absence of overexpression of the 10 HPV-positive genes for HPV-negative status. Only one specimen, TCGA-CR-6472, which was misclassified as HPV-negative, did not over express any of the 11 genes in the classifier by RNA-Seq. Homozygous deletion of CDKN2A and mutations of TP53 are present, as expected, exclusively in the HPV-negative specimens. Mutations, amplifications and over expression of PIK3CA were observed in both HPV-positive and HPV-negative OPSCC tumors