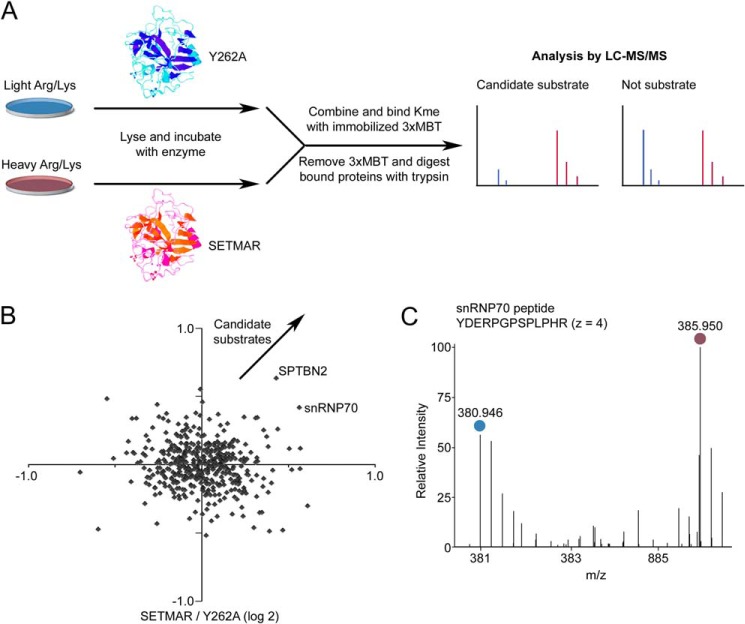
FIGURE 4.
Proteome-wide analysis of proteins methylated by SETMAR in cell lysate identifies snRNP70 as a candidate substrate. A, schematic of the procedure for in vitro labeling of cell lysates prepared with stable isotopic labeling, enrichment of methylated proteins by the immobilized 3xMBT domain, and quantitative analysis by LC-MS/MS. Candidate substrates are identified by the their enzyme-dependent association with 3xMBT. B, relative enrichment of proteins bound by 3xMBT from lysate methylated by GST-SETMAR compared with the Y262A mutant as a negative control. The axes show log base-2 of relative enrichment, and each axis represents an independent experiment with isotopic labels reversed. Positive values are defined so that candidate substrates are in the top right quadrant. C, an example mass spectrum for a peptide from snRNP70 showing the amount bound by 3xMBT from lysates treated with SETMAR or inactive enzyme. The blue circle indicates peptide from light SILAC lysate (treated with the Y262A mutant), and purple indicates heavy SILAC lysate (treated with active SETMAR).