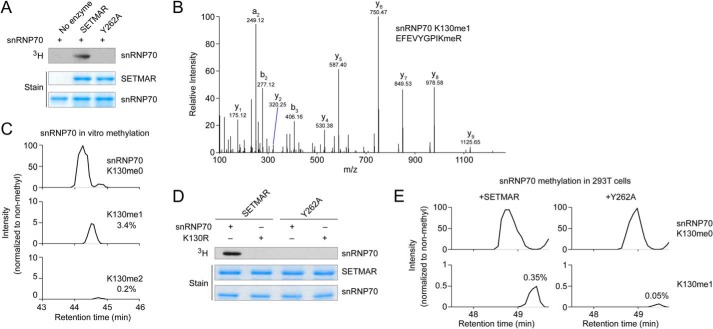
FIGURE 5.
SETMAR methylates snRNP70 at Lys-130. A, radiolabeling assay with GST-SETMAR or the Y262A mutant and snRNP70 (residues 1–204). B, fragmentation spectrum showing in vitro monomethylation of snRNP70 Lys-130 by SETMAR in vitro using deuterated SAM and digested with trypsin. The Orbitrap mass analyzer was used to obtain high-resolution mass accuracy. C, HPLC elution profiles for non-methyl, monomethyl, and dimethyl snRNP70 Lys-130 from tryptic digestion after in vitro methylation by GST-SETMAR using deuterated AdoMet. HPLC elution profiles show a 10-ppm mass window around expected peptide masses (peptide sequence EFEVYGPIKR, Lys-130 is underlined; m/z are 619.332, 628.850, and 636.348). D, radiolabeling of wild-type or K130R mutant snRNP70 (1–204) by GST-SETMAR or the Y262A catalytically inactive mutant. E, snRNP70 was expressed in HEK293T cells with N-terminal 3xFLAG and either active SETMAR or the Y262A mutant. snRNP70 was enriched using FLAG M2 antibody, and methylation at Lys-130 was analyzed by in-gel tryptic digestion and LC-MS/MS. HPLC chromatograms show a 10 ppm mass window around expected peptide masses (the same peptide as in C, methyl mass 14.0157 Da; m/z are 619.332 and 626.340).