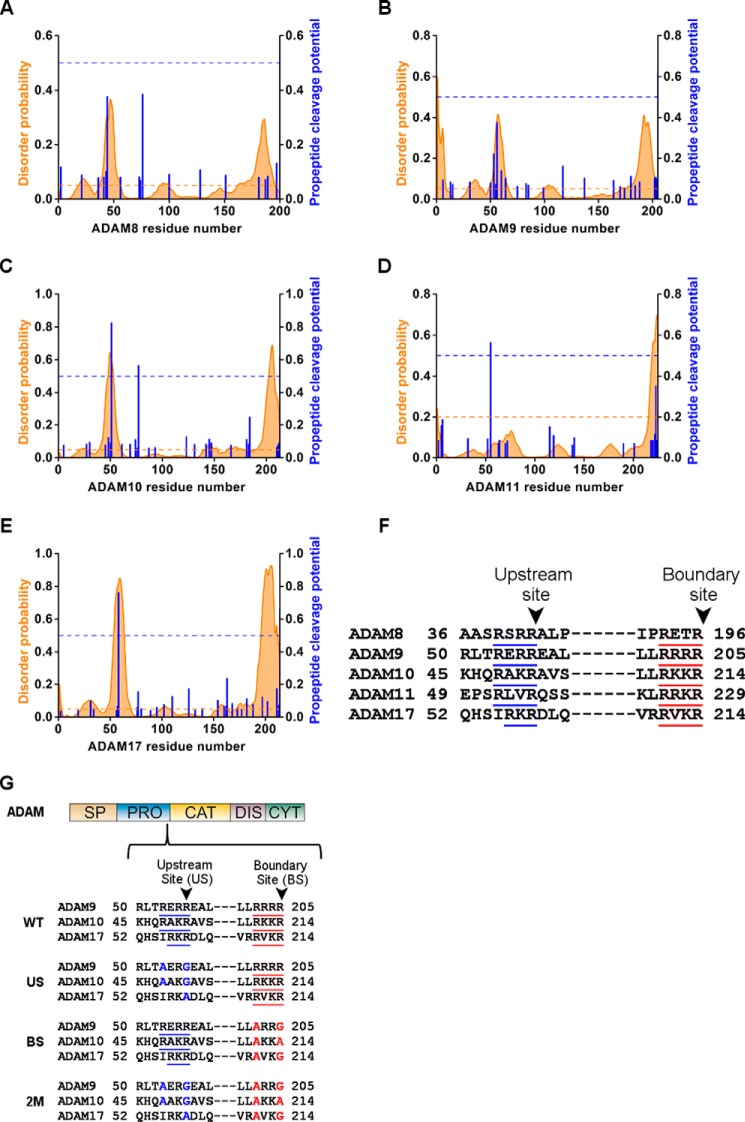
FIGURE 1.
ADAM pro-domain sequence alignment of the potential PC sites. Disordered probability predicted by Disopred server (orange graph), with a threshold of 5% (orange dashed line), and propeptide cleavage potential as predicted by ProP server (blue column), with a threshold of 0.5 (blue dashed line) in the pro-domain sequence of ADAM8 (A), ADAM9 (B), ADAM10 (C), ADAM11 (D), and ADAM17 (E). Sequence alignment of the pro-domain of ADAM8, -9, -10, -11, and -17 in the predicted PC sites (F). Schematic presentation of the ADAM domains structure; SP, signal peptide; PRO, pro-domain; CAT, catalytic domain; DIS, disintegrin domain; CYT, cytoplasmic tail, and below the detailed sequence where the blue underline marks the predicted upstream PC site and red underline marks the boundary site (G). The residues marked in blue are the mutations introduced in the upstream PC sites and in red are the mutations in the boundary sites of ADAM9, -10, or -17 in the US, BS, and 2M mutants.