FIGURE 8.
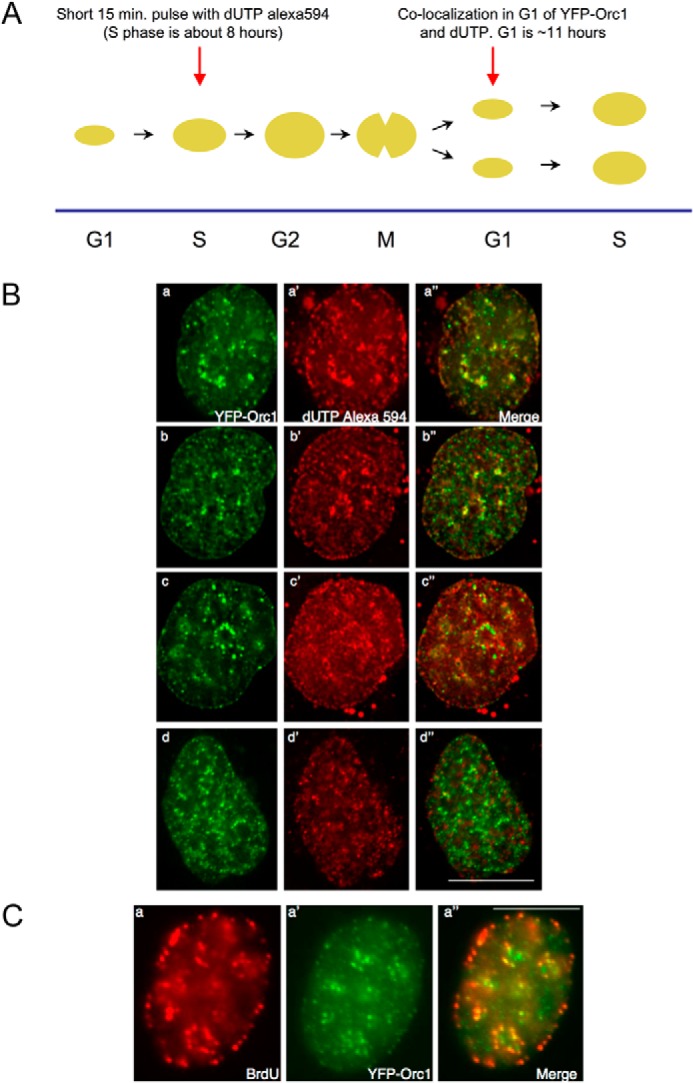
Orc1 localization in G1 phase overlaps with an inherited spatiotemporal pattern of DNA replication. A, scheme for the experiment. Human U2OS cells expressing YFP-Orc1 (green) were transfected for 15 min with dUTP Alexa 594 (red) (see “Experimental Procedures”). Following washing out the label and incubating for ∼12–14 h (ensuring the cells progress from S phase in generation 1 to G1 of generation 2), live cells were imaged every 5 min using a Delta Vision optical sectioning deconvolution instrument (Applied Precision) on a Olympus microscope with a ×63 1.4NA objective. B, top two panels represent significant overlap of YFP-Orc1 with the dUTP Alexa 594, both in nuclear and nucleolar periphery (merge is panels a and b). YFP-Orc1 and dUTP Alexa 594 do not show any overlap but are immediately adjacent to each other (panels c–c″), and (panels d–d″) show the expected most common situation of no overlap between YFP-Orc1 and dUTP Alexa 594. Scale bar denotes 5 μm. Overlap was seen in 6% of all YFP-Orc1-positive cells. C, cells expressing YFP-Orc1 were incubated for 10 min with BrdU, following which BrdU was extensively washed out. Following ∼12–14 h incubation at 37 °C (ensuring the cells enter from S phase in generation 1 to G1 of generation 2), cells were fixed in 2% formaldehyde. BrdU was detected using anti-BrdU mouse mAb (red) in YFP-Orc1 (green)-positive cells. Orc1 was observed both in the nuclear and nucleolar periphery. In a population of cells Orc1 showed significant overlap with BrdU-positive regions. Scale bar denotes 5 μm.
