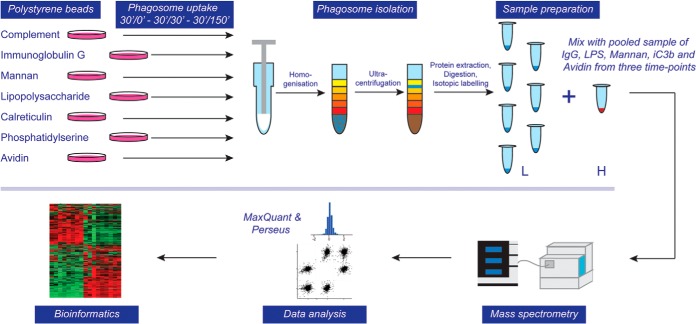
Fig. 2.
Flow diagram of experimental approach for proteomic analysis. Phagosomes were formed by inoculating murine bone marrow-derived macrophages (BMDMs) with polystyrene beads coated with various ligands for 30 min and a subsequent chase for 0, 30 or 150 min. Cells were homogenized and phagosomes isolated through ultracentrifugation in a sucrose gradient. Phagosomal proteins were extracted, digested, and isotopically labeled (light), whereas a mixed pool of phagosome samples that serve as a control was labeled with heavy isotopes. Combined samples were analyzed by LC-MS/MS on an Orbitrap Velos Pro mass spectrometer, data analysis was performed using MaxQuant and Perseus software suites and bioinformatic and statistical analyses were performed using in-house tools.