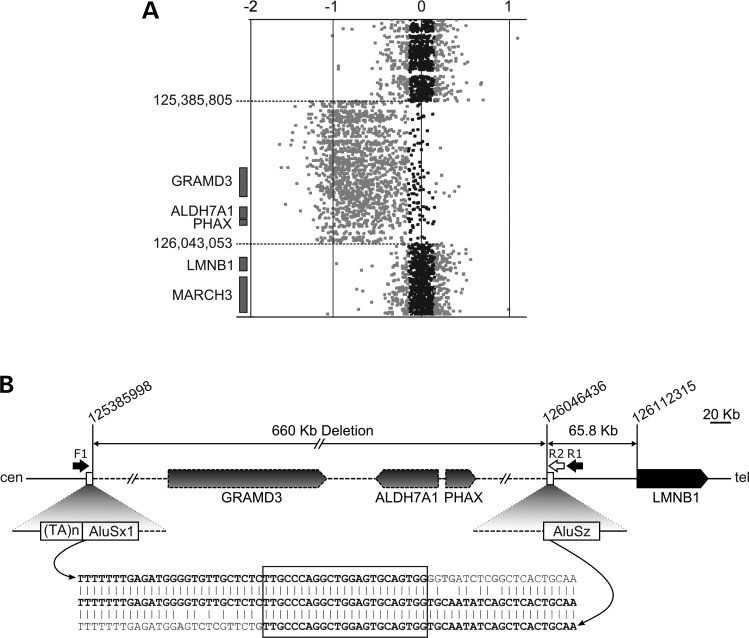
Figure 2.
Characterization of a large deletion 66 kb upstream of LMNB1. (A) Custom a-CGH graphical output. Values on top represent the log ratio of the probes (log2 intensity of Cy5/Cy3 fluorochromes): expected values are from −0.7 to −1 for a deletion, 0 (zero) for normal and 0.5–1 for a genomic duplication. The position of the first normal probe is reported on the left, along with a schematic representation of the genes involved (gray rectangles). (B) Schematic of the deleted genomic region in which the deleted genes have dashed border and the arrowhead shows the transcription direction. The position of the primers used to amplify and sequence the breakpoint (not in scale) is shown (black and white arrows; F1, R1 and R2). The two Alu elements (AluSx1 and AluSz) and a (TA)n repeat are shown. Below, the sequence of the breakpoint: a stretch of 23 identical bases are shared between the two regions (boxed).