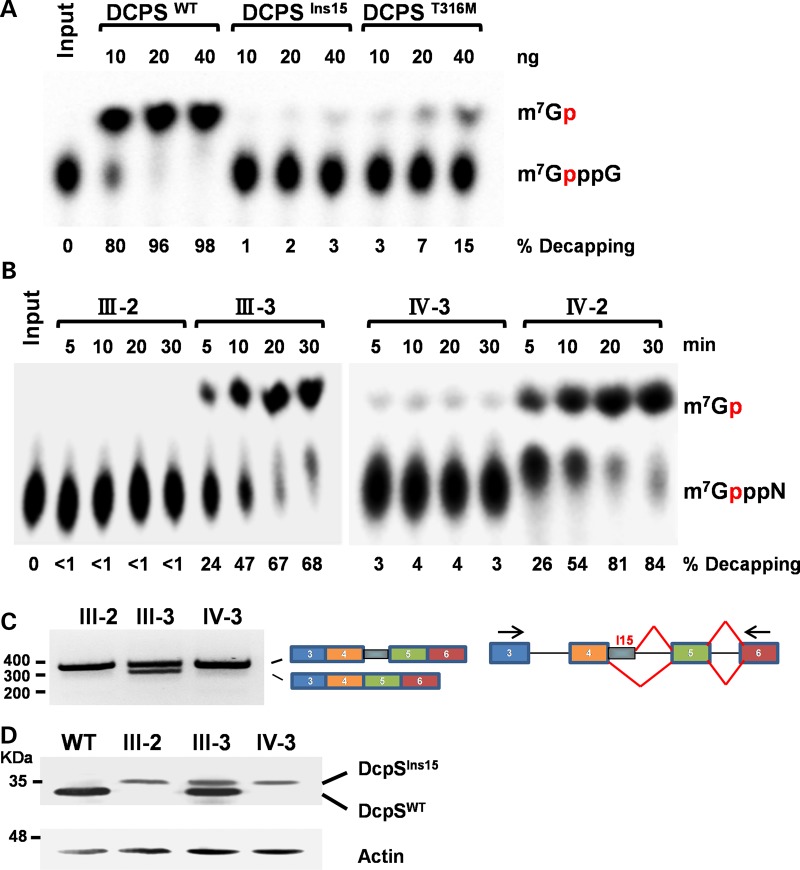
Figure 3.
Functional analysis of DCPS variants. (A) DCPS in vitro decapping assay. mRNA decapping was assayed using recombinant full-length DCPS wild-type (DCPSWT) or altered [splice mutation/45 bp insertion (DCPSIns15) and p.Thr316Met(DCPST316M)] DCPS protein in pET28a (Novagen, EMD Biosciences) vector expressed and purified from Escherichia coli BL21(DE3) cells. Migration of the input cap structure (m7GpppG) or m7GMP product (m7Gp) is designated, with the red ‘p’ denoting the location of the 32P. The percent decapping is indicated at the bottom of the panel. (B) DCPS activity in lymphoblastoid cell lines. Lymphoblastoid cell lines from homozygous affected (III-2 and IV-3) and heterozygous unaffected (III-3 and IV-2) family members were used for decapping assays. Decapping products and labeling are as in (A). (C) RT–PCR analysis for DCPS. RT–PCR across the DCPS splice mutation (from exons 3 to 6) was performed using mRNA from patients (III-2 and IV-3) and unaffected family member (III-3), plus cartoon indicating exon usage. (D) Western Blot of DCPS. Wild-type and variant DCPS protein from the indicated lymphoblast cell lines were detected by western blotting to demonstrate migration of the variant DCPS protein in homozygous and heterozygous individuals. A wild-type lymphoblastoid cell line is shown to indicate the position of wild-type DCPS.