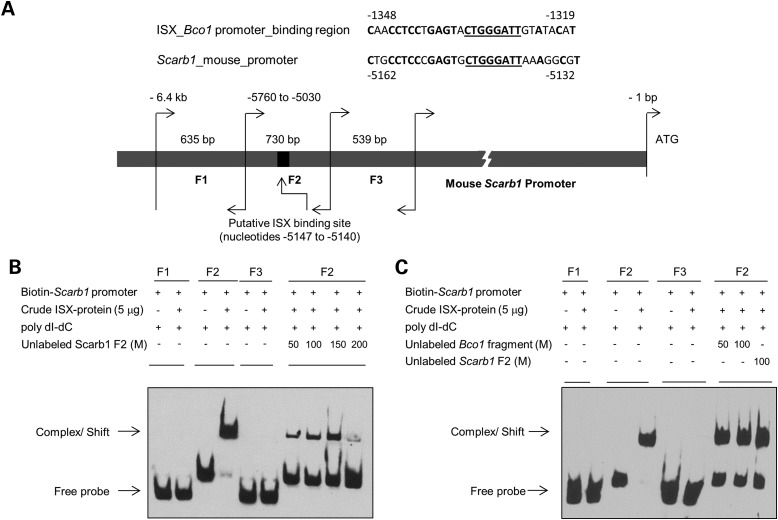
Figure 3.
ISX protein binds to the SCARB1 promoter fragment. (A) The mouse SCARB1 gene is flanked by a putative ISX-binding site, which shows ∼70% sequence homology to the previously identified ISX-binding site upstream if the BCO1 gene. Nucleotide sequence similarities between the putative ISX-binding sites upstream of the BCO1 and SCARB1 genes are underlined. Genomic DNA was isolated from mouse intestine and using primer pairs listed in Materials and Methods. Three overlapping fragments (F1–F3) were amplified as indicated in Materials and Method. Individual DNA fragment sizes ranged from ∼540 to 730 bp. The 8-bp core ISX-binding nucleotide sequence within fragment 2 (−5147 to −5140) is underlined. Individual SCARB1 fragment sizes and positions are given relevant to the SCARB1 ATG start site. (B and C), in EMSA, individual biotin-labeled SCARB1 fragments were incubated with supernatant protein fraction of BL21 cells that expressed recombinant ISX. SCARB1 fragment F2 shifted in the presence of protein extracts containing ISX protein as indicated by the arrow. No such shift in electro-mobility was observed with fragments 1 and 3 (F1 and F3). Unlabeled SCARB1 fragment 2 [50–200 molar (M) excess] competed with the ISX binding, as shown in the right most lanes of the EMSA gels. Unlabeled SCARB1 fragment 2 and/ or unlabeled BCO1 fragment containing the previously identified ISX-binding nucleotide sequence competed with ISX binding to labeled SCARB1 fragment 2. M represents molar excess concentrations of unlabeled fragments as indicated. In all EMSAs, biotin-labeled SCARB1 promoter fragments (±ISX protein) were resolved on 6% PAGE gels, and labeled fragments were visualized by chemo-luminescence. The figures shown are representatives of images from at least two independent experiments using different ISX protein crude extract preparations.