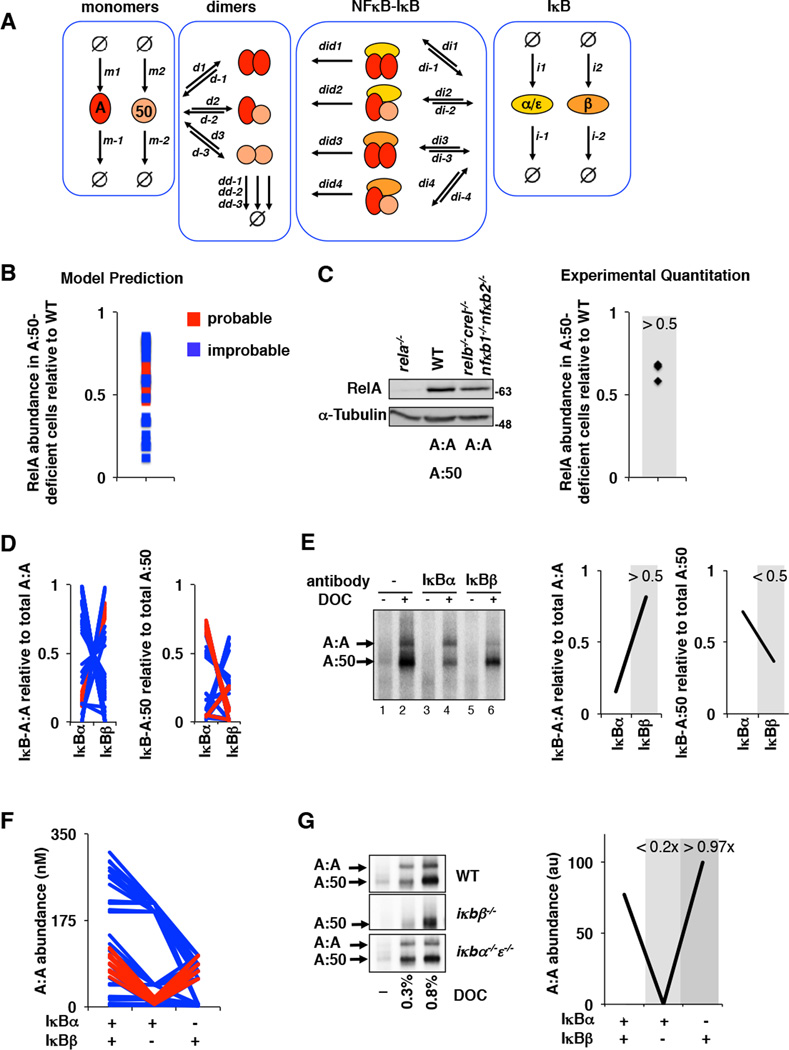
Figure 5. Iterative testing and refinement of the IκBβ -RelA homodimer preference model.
A. A model schematic of the Rel-NFκB dimer generation module, as in Figure 3A but with two IκBs, IκBα and IκBβ, necessitating i1 and i2 synthesis rate constants, i−1 and i−2 degradation rate constants, di1,di2, di3, and di4 IκB-NFκB association rate constants. di−1, di−2, di−3, and di−4 IκB-NFκB dissociation rate constants, and did1, did2, did3, and did4 IκB-NFκB degradation rate constants of IκB to release NFκB.
B. Computational prediction of RelA protein abundance in A:50-deficient cells (relative to wild type cells), using probable (red) and improbable (blue) affinities shown in Figure 4B and listed in Supplementary Table 5.
C. Immunoblot against RelA of whole cell extracts from RelA-deficient, wild type, and RelA-only MEFs. The right panel shows the quantitation of three independent experiments. These consistently showed values of > 0.5.
D. Computational predictions of the abundances of NFκB dimers RelA:RelA (left) and RelA:p50 (right) bound to either IκBα or IκBβ as indicated. These calculations used probable (red) and improbable affinities (blue) as in panel B.
E. Electrophoretic mobility shift assay of deoxycholate (DOC)-treated cytoplasmic extracts (which results in the separation of NFκB dimers from IκBs) to quantitate IκB-bound NFκB dimer abundance. Prior immunodepletion of the extracts with antibodies against IκBα or IκBβ allows quantitation of NFκB bound to the remaining IκB isoform. Data shown is representative of three independent experiments, and the right panel shows the corresponding quantitation.
F. Computational predictions of the basal abundances of the RelA:RelA homodimer in wild type, IκBβ -deficient, and IκBα-deficient MEFs using probable (red) and improbable (blue) affinities as in panel B.
G. Electrophoretic mobility shift assays (left panel) of DOC-treated cytoplasmic extracts of wild type, IκBβ -deficient, and IκBα/ε-deficient MEFs. Shown is a representative result of three independent experiments, and the right panel shows the corresponding quantitation.