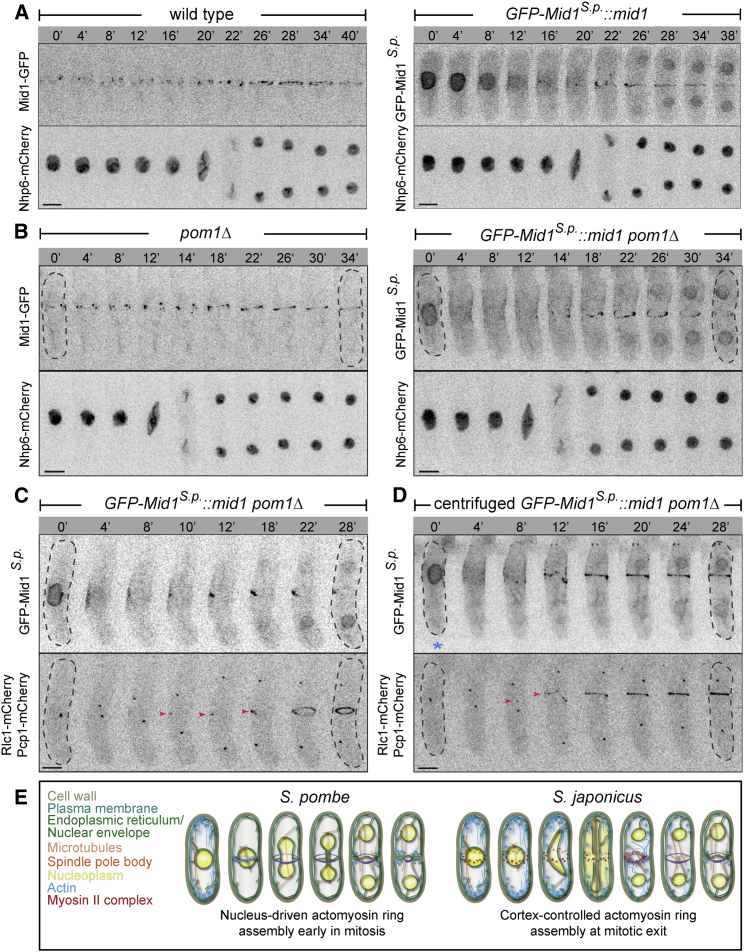
Figure 4.
Replacement of S. japonicus Mid1 with Its S. pombe Ortholog Is Sufficient to Reconstruct Nucleus-Driven Division Site Positioning
(A) Time-lapse maximum-projection images of S. japonicus cells co-expressing the endogenous GFP-tagged Mid1 (left) or its S. pombe ortholog GFP-Mid1S.p. (right) together with the nuclear marker Nhp6-mCherry.
(B) Time-lapse maximum-projection images of pom1Δ S. japonicus cells co-expressing either Mid1-GFP (left) or GFP-Mid1S.p. (right) and Nhp6-mCherry.
(C) Time-lapse maximum-projection images of pom1Δ S. japonicus cells co-expressing GFP-Mid1S.p. with Rlc1-mCherry and the SPB marker Pcp1-mCherry to indicate mitotic progression. Dashed lines indicate cell boundaries. Red arrowheads indicate Rlc1-mCherry signal at the equatorial cortex.
(D) Time-lapse maximum-projection images of pom1Δ S. japonicus cell co-expressing GFP-Mid1S.p., Rlc1-mCherry and Pcp1-mCherry, where the nucleus was displaced toward a non-growing end by centrifugation. Dashed lines indicate cell boundaries. Red arrowheads indicate Rlc1-mCherry signal at the cortex. Blue asterisk indicates a growing cell tip.
(E) Diagrams summarizing the modes of cytokinesis in S. pombe and S. japonicus.
For (A)–(D), time is in minutes. Scale bars represent 5 μm. See also Figure S4.