Figure 1.
Basket Tethering to the NPC via the Nup1 and Nup60 N Termini
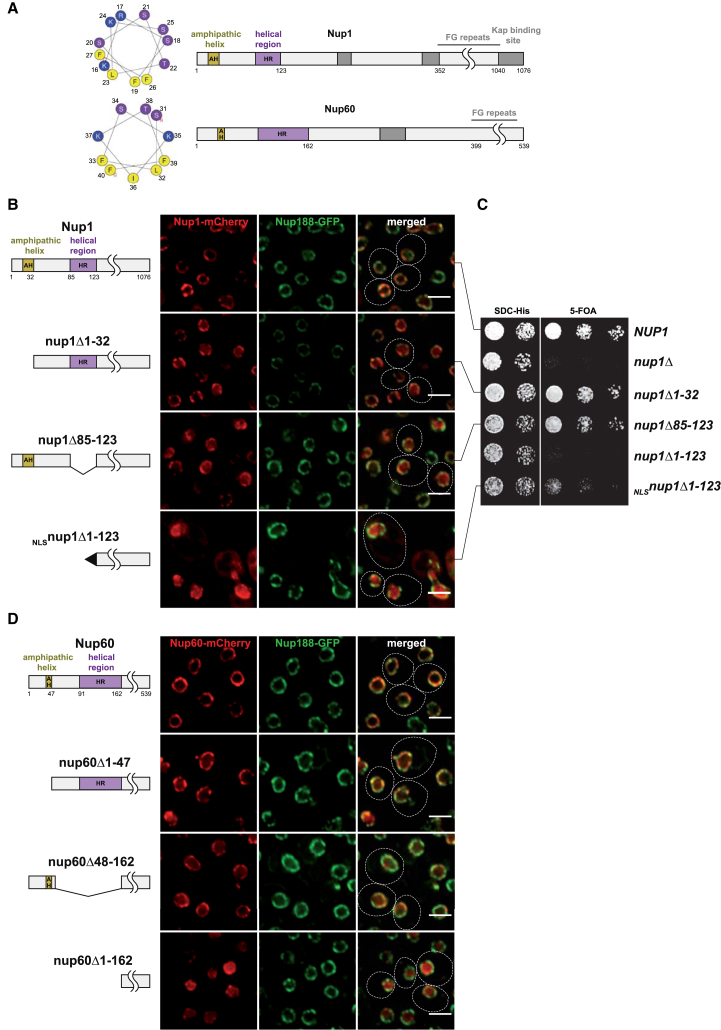
(A) Nup1 and Nup60 motif organization based on secondary structure predictions and sequence conservation (https://www.predictprotein.org/). Predicted amphipathic helices (AH; yellow) and α-helical regions (HR; purple) are highlighted. Helical wheel projections (http://heliquest.ipmc.cnrs.fr/) of the Nup1 (16-27) and Nup60 (31-40) AHs. Boundaries are drawn to scale, gray boxes indicate other motifs.
(B) Double-fluorescence microscopy of live nup1Δ cells expressing genomically tagged Nup188-GFP and plasmid-borne NUP1 alleles (endogenous promoter) with a C-terminal mCherry tag. Note that NLSnup1Δ1-123 cells are frequently enlarged. NLS, SV40 large T-antigen NLS. Dashed lines mark the contour of cells. Scale bar represents 3 μm.
(C) Growth analysis of the NUP1 alleles used in (B). A nup1Δ strain harboring wild-type NUP1 (URA plasmid) was transformed with wild-type NUP1, empty vector, or the indicated nup1 alleles (HIS plasmids). Growth was followed on SDC-His (loading control) and on SDC+5-fluoroorotic acid (5-FOA) plates to shuffle out the URA cover plasmid. Cells were spotted onto plates in 10-fold serial dilutions and incubated for 2 (SDC-His) or 3 days (5-FOA) at 30°C.
(D) Double-fluorescence microscopy of live nup60Δ cells expressing genomically tagged Nup188-GFP and plasmid-borne NUP60 alleles with a C-terminal mCherry tag (endogenous promoter). Scale bar represents 3 μm.