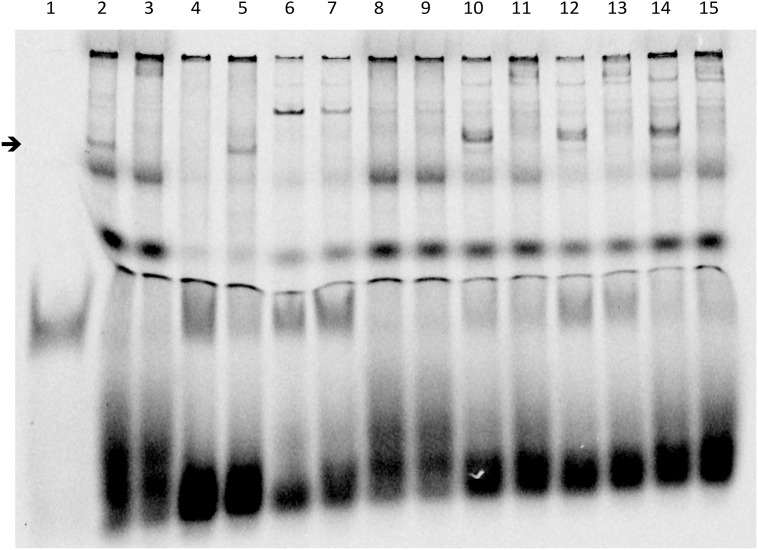
Fig 3. Electro Mobility Shift Assay Experiments.
EMSA was performed with the alphaGSU-luego2Cy5 probe, corresponding to a consensus binding sequence of LHX4 identified on the alphaGSU murine promoter (5'-gtgccctggtctgg ggtacttagctaattaaatgtg-3 ‘; 3'-cacgggaccagacc-ccatgaatcgattaatttacac-5'). Supershift was performed with a monoclonal anti-myc antibody. Arrow indicates position of the expected LHX4 DNA binding. Lanes are identified as follows: 1, free probe; 2, LHX4 wt (band corresponding to LHX4 identified by the arrow); 3, LHX4 wt + anti-myc antibody (supershift, no LHX4 band visualized at the arrow level); 4, non mutated competitor (LHX4 band is not visualized, as the probe is bound by the competitor); 5, mutated competitor (LHX4 band is visualized at the arrow level as the competitor can not bind the probe); 6, PcDNA3.1 (empty vector); 7, PcDNA3.1 + anti-myc antibody; 8, W204X; 9, W204X + anti-myc antibody (supershift); 10, DelK242; 11, DelK242 + anti-myc antibody (supershift); 12, N271S; 13, N271S + anti-myc antibody (supershift); 14, Q346R; 15, Q346R + anti-myc antibody (supershift).