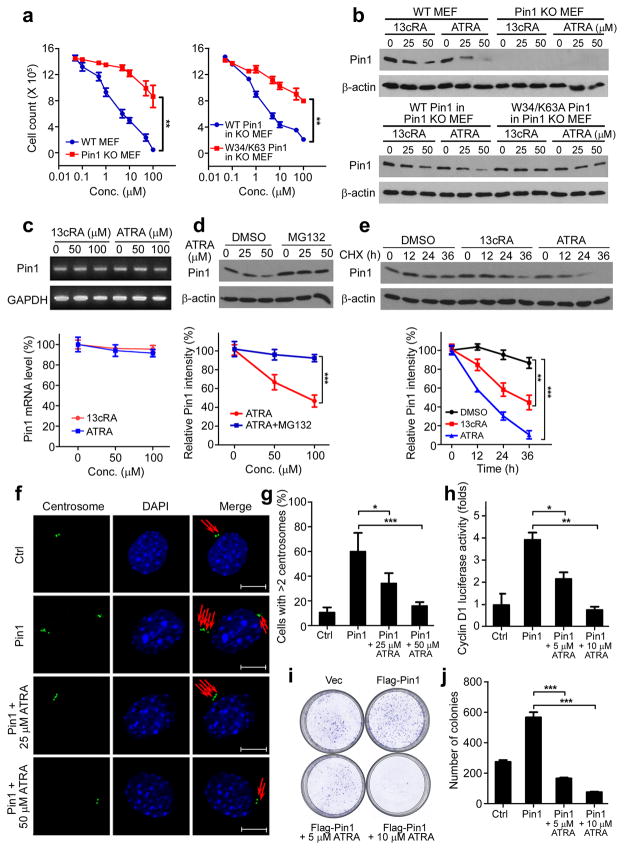
Figure 2. ATRA causes Pin1 degradation and inhibits its oncogenic function in cells.
(a) WT and Pin1 KO MEFs (left) or Pin1 KO MEFs reconstituted with WT- or W34/K63A-Pin1 (right) were treated with indicated concentrations of ATRA for 72 h, followed by MTT cell growth assay with the readout absorbance measured by 570 nm (mean ± s.d. of three experiments)
(b) WT and Pin1 KO MEFs (upper) or Pin1 KO MEFs reconstituted with WT- or W34/K63A-Pin1 (lower) were treated with the indicated concentrations of13cRA or ATRA for 72 h. Pin1 abundance was assessed by immunoblotting (mean ± s.d. of three experiments).
(c) MEFs were treated with the indicated concentrations of ATRA or 13cRA for 72h, followed by quantitative RT-PCR to detect Pin1 mRNA. Top panel shows representative gel, and bottom graph shows quantification (mean ± s.d. of three experiments).
(d) MEFs were treated with the indicated concentrations of ATRA for 48 h followed by co-treatment of 10 μM MG132 or DMSO control for additional 24 h before harvest. Pin1 abundance was measured by immunoblotting. Top panel shows representative gel, and bottom graph shows quantification (mean ± s.d. of three experiments).
(e) MEFs were treated with 50 μM ATRA, 50 μM 13cRA or DMSO control for 24h, followed by cycloheximide (CHX) chase for the indicated times. Pin1 abundance was measured by immunoblotting. Top panel shows representative gel, and bottom graph shows quantification (mean ± s.d. of three experiments).
(f and g) NIH3T3 cells stably expressing Flag-tagged Pin1 or empty vector (Ctrl) were treated with indicated concentrations of ATRA for 72 h, followed by immunostaining with antibodies specific for γ-tubulin to detect centrosomes (red arrow). Scale bar, 10μM. Cells containing over 2 centrosomes were quantified from 3 independent experiments with over 100 cells in each (g).
(h) SKBR3 cells were co-transfected with a cyclin D1 promoter-driven luciferase construct, and with Flag-Pin1 or control vector. Cells were treated with indicated concentrations of ATRA for 72 h. Luciferase activity was measured by reporter gene assay (mean ± s.d. of three experiments).
(i and j) SKBR3 cells were co-transfected with Flag-Pin1 or empty vector, and were then treated with indicated concentrations of ATRAfor 48 h. Foci formation assay was applied. Representative image (i), foci quantification in (j) (mean ± s.d. of three experiments).
*P < 0.05, **P < 0.01, ***P < 0.001, as determined by Student’s t-test.