Figure 1. ChIRP-MS method and validation.
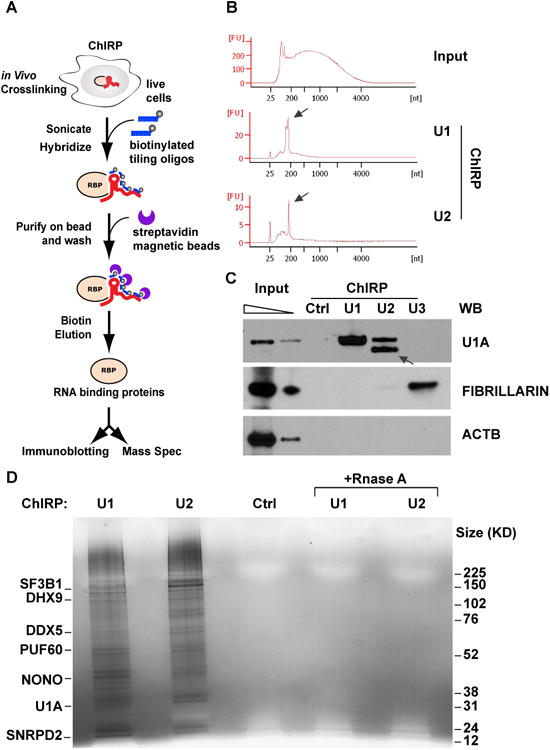
(A) outline of the ChIRP-MS workflow. Briefly, RNP complexes are crosslinked in vivo by 3% formaldehyde for 30min, and solubilized by sonication. Target ncRNA are pulled out by biotinylated anti-sense oligos, and associated proteins are eluted with free biotin, separated by electrophoresis, and each size fraction is subjected to LC/MS-MS identification. (B) Distribution of input, and U1 and U2-enriched RNA sizes, as determined by Bioanalyzer (Agilent). (C) Proteins retrieved by U1, U2, U3 and control probes, analyzed by immunoblotting. Arrow indicates the U1A close homolog, U2B, cross-identified by U1A antibody. (D) Proteins retrieved by U1, U2, Rnase-treated controls and non-targeting probe control, visualized by silver staining. Major proteins enriched are indicated on the left.
