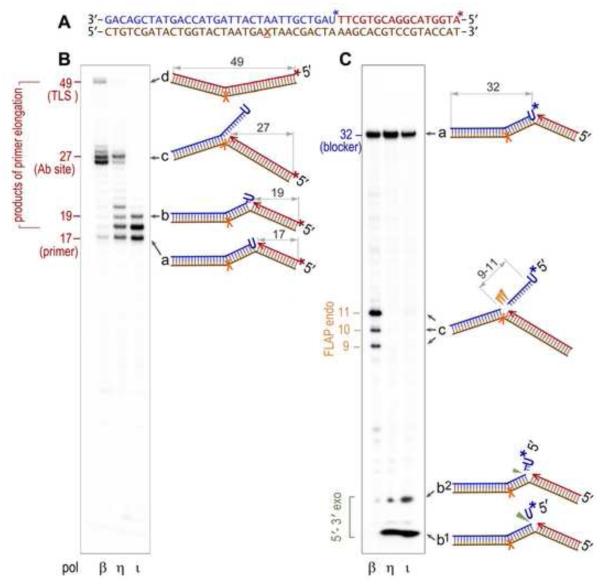
Fig. 2. Processing of a nicked DNA substrate with a single ribonucleotide (U) in the 5' terminus of the nick by the combined action of a DNA polymerase and nuclease.
A. Sequence of the oligonucleotides used to generate the nicked DNA substrate (also shown schematically to the right of each gel panel as “a”), consisting of the 49-mer DNA template (brown) with an abasic site (X) annealed with a 17-mer primer (red) and a 32-mer downstream blocker (blue). The template in which either primer, or blocker, is P32-labeled (indicated by the star, *), together with the purified recombinant human DNA polymerase (pol β, pol η, or pol ι) and Fen1 protein was used in in vitro reactions B. P32-labeling of the 17-mer primer (red *) allows for visualization of the primer extension products. The processivity of the strand displacement synthesis, decreasing in the order pol β> pol η> or pol τ□ can be judged by the intensity of the intermediate bands such as the 19-mer oligonucleotide, whose structure (“b”) is schematically shown to the right of the gel. The ~27 nt-long bands correspond to replication blockage by the abasic site on the DNA template (c), and the 49-mer band corresponds to the full-sized product generated by translesion replication to the end of the template (d). C. P32- labeling of the 32-mer blocker (blue *) allows one to visualize products of the 5'→3' exo-(green ▼), and flap endonucleolytic (orange ▼) cleavage.