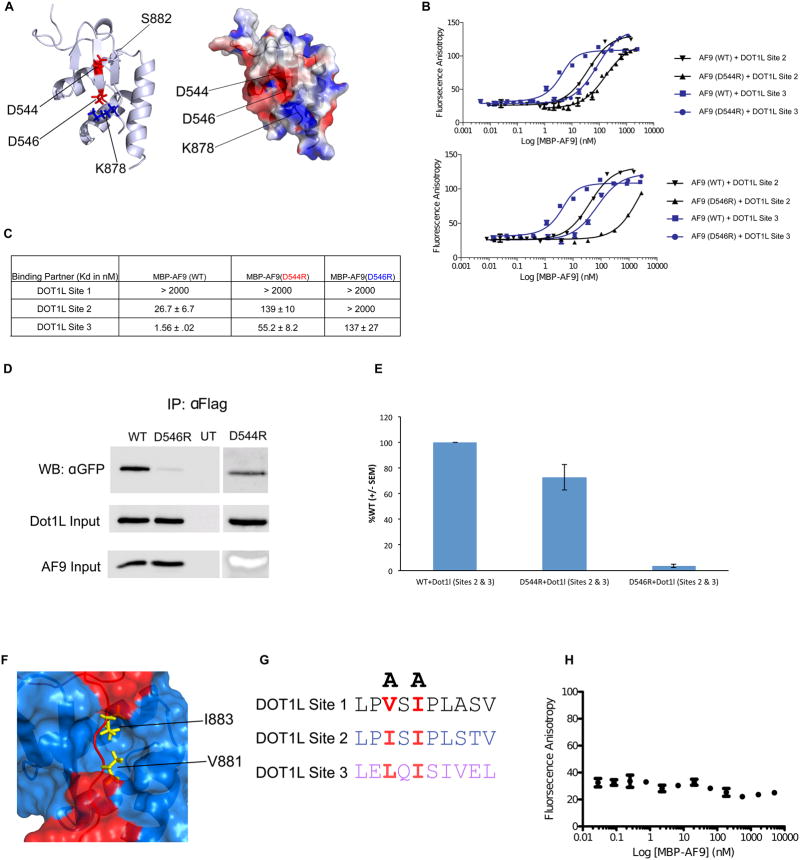
Figure 3. Identification of mutations that disrupt the DOT1L-AF9 interactions.
A) Left: Cartoon representation of the DOT1L-AF9 structure showing D544 and D546, located on the AF9 β hairpin, that make direct interactions with DOT1L. Right: Electrostatic surface representation depicting that D546 (AF9) and K878 (DOT1L) make a charge-charge interaction. B) Results of fluorescence polarization assays for determination of the Kd values for binding of MBP-AF9 AHD (WT), MBP-AF9 AHD (D544R) (Top), and MBP-AF9 AHD (D546R) (Bottom), to DOT1L binding motifs 2 and 3. C) Table of Kd values for binding of MBP-AF9 AHD (WT), MBP-AF9 AHD (D544R), and MBP-AF9 AHD (D546R) to the DOT1L binding motifs. MBP-AF9 (D546R) significantly affects binding of both DOT1L high binding sites 2 and 3, whereas MBP-AF9 (D544R) has a more significant effect only on Site 3. D) and E) Co-IP data of D546R and D544R with the high affinity sites of DOT1L. F) Surface representation of the DOT1L (red) – AF9 (blue) complex. Sidechains of two buried hydrophobic residues form DOT1L, V881 and I883, are shown in yellow. G) Two alanine mutations of similarly positioned hydrophobic residues within each DOT1L binding site used to disrupt AF9 binding. H) Results of fluorescence polarization assay for MBP-AF9 AHD titrated into fluorescently tagged DOT1L peptide with (V881A, I883A), showing no binding.