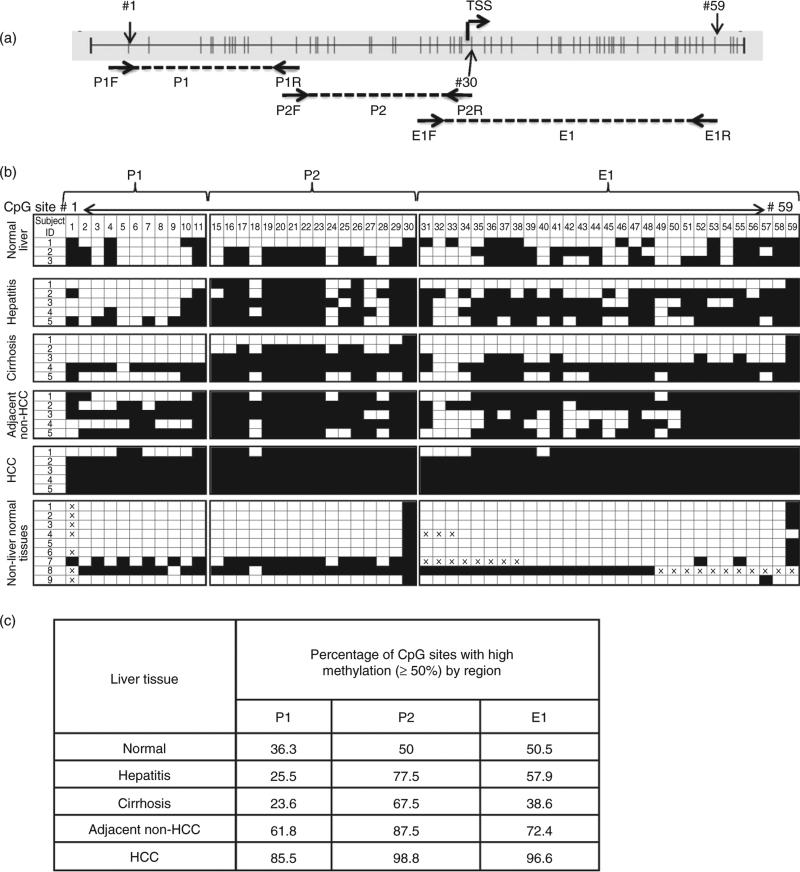
Figure 1.
Bisulfite (BS) sequencing analysis of the RASSF1A promoter and first exon region in normal liver, diseased liver and non-liver normal tissues. (a) RASSF1A promoter region (GenBank accession no. DQ444319.1) and position of BS sequencing primers for three regions: P1 (nucleotides 357–548), P2 (nucleotides 530–736) and E1 (nucleotides 680–981). Vertical lines represent CpG sites; the transcription start site (TSS) is also indicated. CpG sites designated 1–59 are indicated with arrows. Note, the BS sequencing data for CpG sites 12–14 were not available due to sequencing-related technical issues; thus, they are not listed. (b) The extent of DNA methylation in three regions, P1, P2 and E1. Qualitative index of each CpG site (open boxes, <50% methylation detected; filled boxes, 350% methylation detected) is based on the relative heights of cytosine and thymine peaks within the chromatogram generated by a reconstituted standard as shown in the Figure S1. The non-liver normal tissues are: 1, spleen; 2, lung; 3, breast; 4, stomach; 5, colon; 6, trigeminal ganglion; 7, pancreas; 8, kidney; and 9, fetal liver. (c) The percentage of CpG sites with high (350%) methylation per total number of CpG sites examined in each region, P1, P2 and E1, in each liver tissue type as listed in the table. HCC, hepatocellular carcinoma.