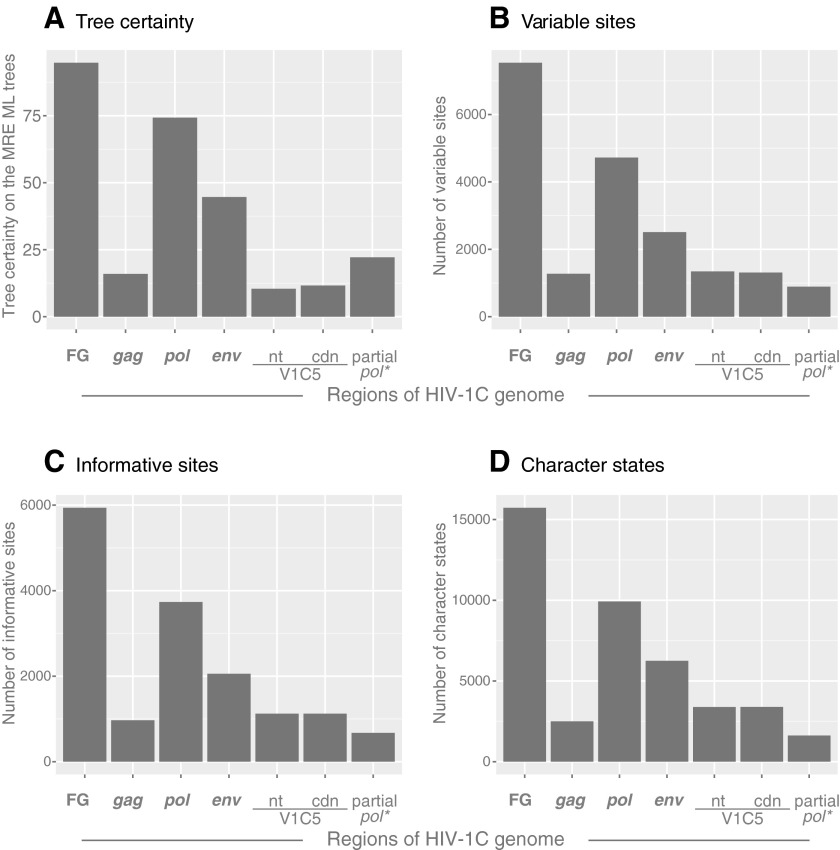
FIG. 2.
Tree certainty, variable and informative sites, and character states. Axis x shows targeted regions across the HIV-1C genome. (A) Relative tree certainty. Internode certainty was quantified by considering the two most prevalent conflicting bipartitions and calculating the log magnitude of their difference. Tree certainty was quantified as the sum of internode certainty over all internodes in a phylogeny.37 Tree certainty scores were calculated in RAxML ver. 8.0.0.52 Axis y shows the relative tree certainty estimated on the extended maximum rule consensus tree. (B) Variable sites. The number of variable sites was computed in MEGA6,47 and is shown on axis y. (C) Informative sites. The number of parsimony-informative sites was computed in MEGA6,47 and is shown on axis y. (D) Character states. The number of character states was computed as the minimum number of parsimony-informative character-state changes, Δmin, according to Wortley and Scotland,50 and is shown on axis y.