FIG. 3.
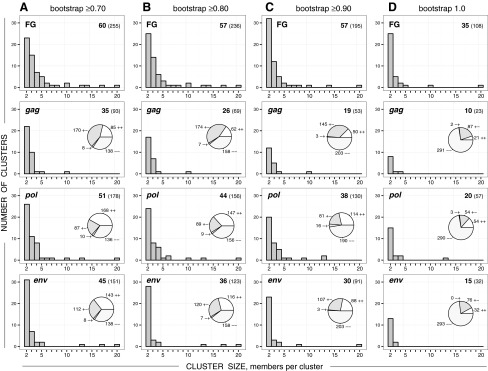
Cluster size distribution in the near full-length HIV-1C genome sequences (FG), gag, pol, and env. Histograms in each graph show the number of clusters (axis y) of specified cluster size (axis x). Numbers in the upper right corner of each graph indicate the number of identified clusters (shown in bold) and the number of viral sequences in clusters (shown in brackets). Pie charts within gag, pol, and env graphs show concordant (++ and −−) or discordant (+− and −+) clustering between near full-length genome (FG) clustering and the corresponding subgenomic region. Column (A): HIV clustering at bootstrap threshold ≥0.70. Column (B): HIV clustering at bootstrap threshold ≥0.80. Column (C): HIV clustering at bootstrap threshold ≥0.90. Column (D): HIV clustering at bootstrap threshold 1.0.
