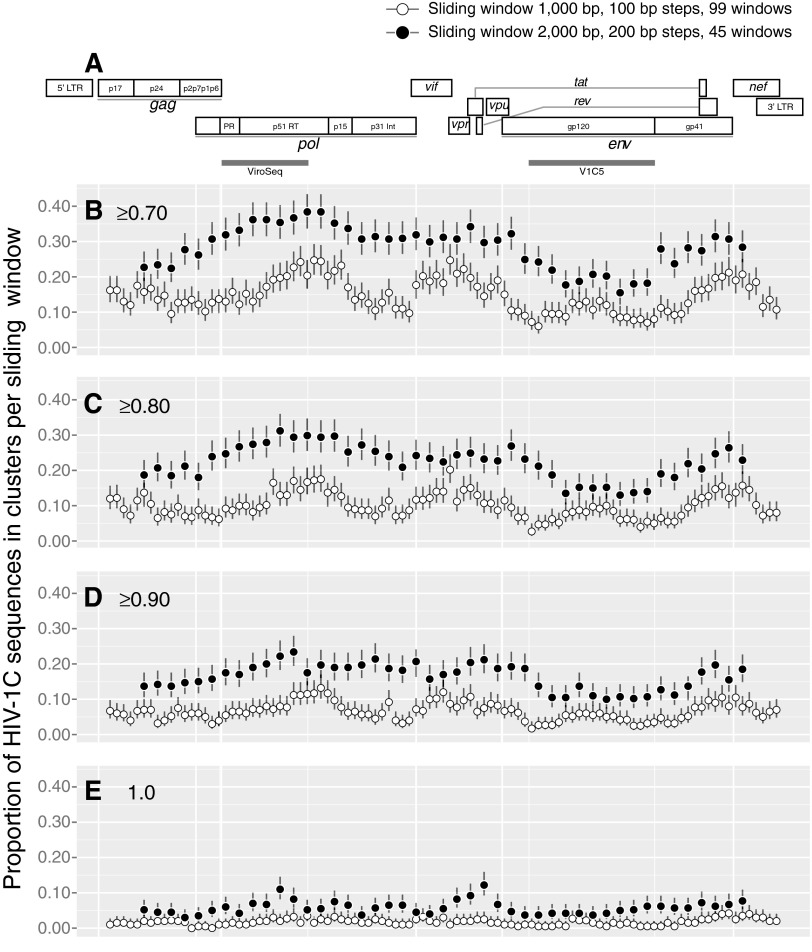
FIG. 4.
Sliding window analysis across the HIV-1C genome. (A) HIV-1 genome structure. The map is drawn according to the multiple sequence alignment of the near full-length genome sequences. gray bars spanning the protease and partial reverse transcriptase sequence in pol and the partial gp120 sequence represent the ViroSeq and V1C5 regions, respectively. (B–E) The extent of HIV-1C clustering for each sliding window at different bootstrap thresholds for cluster identification: (B) ≥0.70, (C) ≥0.80, (D) ≥0.90, and (E)=1.0. A total of 99 sliding windows with a length of 1,000-bp and 100-bp steps are shown as open circles. A total of 45 sliding windows with a length of 2,000-bp and 200-bp steps are presented as filled circles. Bars above and below the circles indicate 95% CI computed for each sliding window.