Figure 2. G-quadruplex structures in switch RNA.
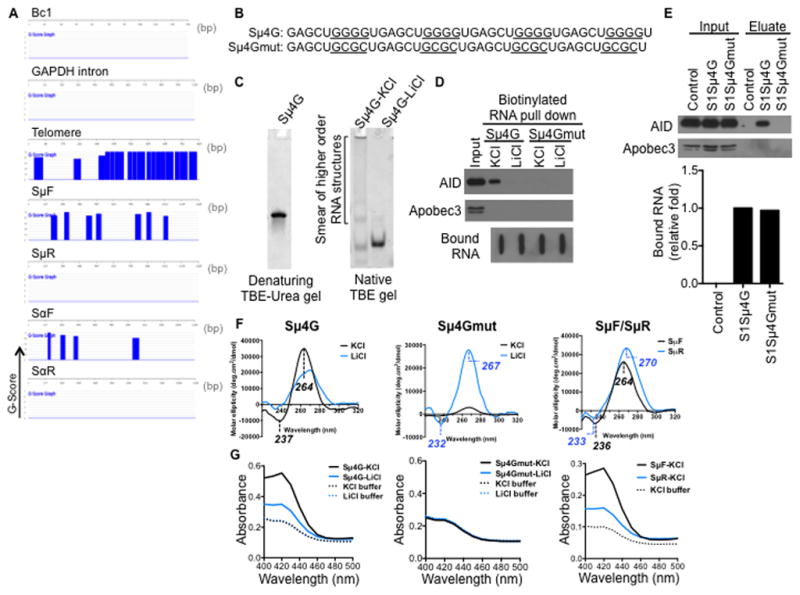
(A) Sense switch RNAs and telomere RNA are predicted to form G-quadruplex structures. QGRS Mapper software was used to assess the G-quadruplex forming potential of the RNA sequences. The probability of G-quadruplex structure formation is reported as a G-score and represented over the corresponding regions in blue.
(B) Sequences of synthetic RNA oligonucleotides. Sμ4G, four Sμ repeats in tandem; mutant Sμ4G (Sμ4Gmut), with G-to-C mutations that abolish guanine tracts. Guanine tracts in Sμ4G and corresponding regions in Sμ4Gmut are underlined.
(C) Sμ4G was resolved on a denaturing gel, or folded in either KCl- or LiCl-containing buffer and resolved on a native gel, and stained with SYBR GOLD following electrophoresis.
(D) Sμ4G associates with AID while Sμ4Gmut does not. RNA pull-down was performed with biotinylated Sμ4G and Sμ4Gmut (folded in either KCl- or LiCl-containing buffer) and stimulated CH12 extracts. Recovered proteins were analyzed by immunoblot and bound RNAs by streptavidin-HRP blot. Result shown is representative of three independent pull-down experiments.
(E) Sμ4G interacts with AID in vivo. CH12 cells stably expressing S1-aptamer tagged Sμ4G or Sμ4Gmut transcripts were stimulated and ribonucleoprotein complexes were isolated on streptavidin beads. CH12 cells expressing anti-sense S1Sμ4G, which is unable to bind streptavidin, were used as control. RNA in the eluates was analyzed by RT-qPCR for amounts of exogenous transcripts relative to S1Sμ4G, while proteins in the eulates were analyzed by immunoblot. Result shown is representative of two independent pull-down experiments.
(F) Circular dichroism (CD) analysis of G-quadruplex structures. CD spectra of Sμ4G, Sμ4Gmut, SμF and SμR RNAs (folded in either KCl- or LiCl-containing buffer). Wavelengths of observed peaks are indicated. Peaks characteristic of parallel G-quadruplexes are (positive- ~260 nm, negative- ~240 nm).
(G) Colorimetric assay of G-quadruplexes. RNAs were folded and incubated with hemin. G-quadruplexes bind hemin and the resultant complex exhibits peroxidase-like activity, which can be detected as an increase in absorbance around 420 nm when substrate is added (Haeusler et al., 2014; Li et al., 2013).
See also Figure S1.
