Table 3. Proportion of fresh organic matter (FOM) derived C to total C and 13C distribution among selected phospholipid fatty acid (PLFA) groups in FOM treated soils on 0 d, 20 d, and 105 d of the incubation.
|
Larch Plantation soil |
Secondary Forest soil |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G+ | G- | Bacteria | Fungi | Actinomycetes | G+ | G- | Bacteria | Fungi | Actinomycetes | ||||||
| Proportion of FOM-derived C to total C in PLFAs (%) | |||||||||||||||
| 20d | |||||||||||||||
| +leaves | 5.37a | 4.96a | 5.22a | 9.66b | 1.80b | 9.67a | 10.70a | 10.03a | 15.69a | 5.87a | |||||
| +stalks | 4.78b | 4.77a | 4.78b | 10.40a | 2.55a | 7.68b | 9.70b | 8.39b | 16.32a | 6.82a | |||||
| 105d | |||||||||||||||
| +leaves | 4.49a | 3.81a | 4.27a | 6.62a | 4.08a | 9.17a | 9.02a | 9.12a | 12.13a | 7.66b | |||||
| +stalks | 4.27a | 3.74a | 4.09a | 7.64a | 4.96a | 9.40a | 9.18a | 9.33a | 12.38a | 9.27a | |||||
| 13C distribution among PLFA groups (%)* | |||||||||||||||
| 20d | |||||||||||||||
| +leaves | 25.39a | 12.67b | 38.06a | 26.96b | 2.29b | 25.12a | 15.09a | 40.20a | 26.19b | 3.91b | |||||
| +stalks | 23.94a | 13.84a | 37.78a | 30.97a | 3.35a | 22.69b | 15.66a | 38.36b | 30.26a | 4.86a | |||||
| 105d | |||||||||||||||
| +leaves | 29.64a | 12.06a | 41.70a | 22.63b | 6.51a | 30.36a | 14.68a | 45.05a | 21.78a | 6.39b | |||||
| +stalks | 27.18b | 12.34a | 39.52a | 26.20a | 8.57a | 30.61a | 14.73a | 45.34a | 22.22a | 8.12a | |||||
Letters in the same column for each sampling time denote significant differences (P < 0.05) between FOM treatments (n = 4). Note: The proportion of FOM-derived C to total C in each microbial group (e.g. bacteria, fungi) was calculated as: 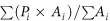 ; 13C distribution proportion in each microbial group (e.g. bacteria, fungi) was calculated as:
; 13C distribution proportion in each microbial group (e.g. bacteria, fungi) was calculated as: 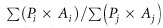 ; where Pi and Ai are the proportion of FOM-derived C to total C and the abundance of each PLFA for a special microbial group (e.g. bacteria, fungi), respectively; while Pj and Aj (j = 1, 2∙∙∙∙∙∙16) are those for all 16 PLFAs detected.
; where Pi and Ai are the proportion of FOM-derived C to total C and the abundance of each PLFA for a special microbial group (e.g. bacteria, fungi), respectively; while Pj and Aj (j = 1, 2∙∙∙∙∙∙16) are those for all 16 PLFAs detected.
*As the results of common saturated PLFAs (16:0, 18:0), which were not assigned to a taxonomic group (bacteria, fungi or actinomycetes), were not shown, the sum of these % was less than 100%.
