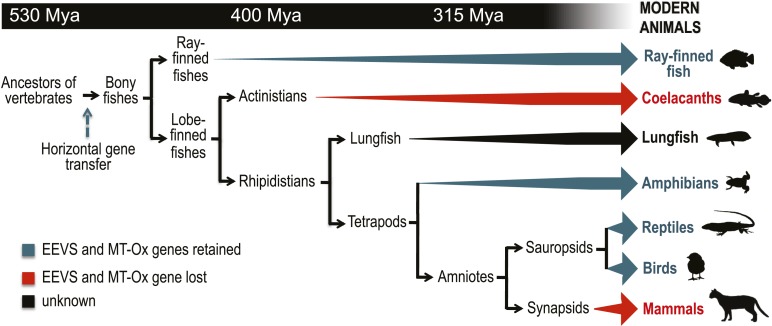
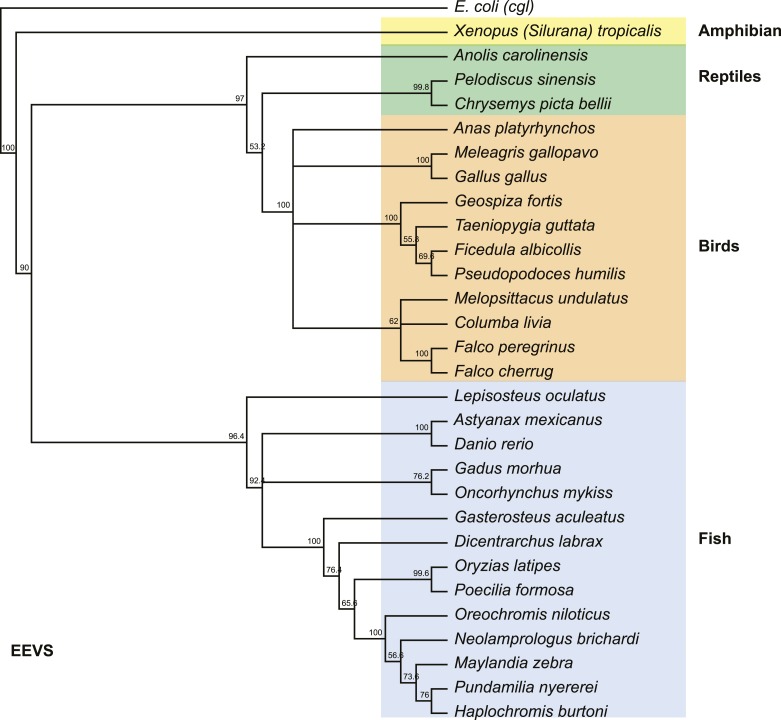
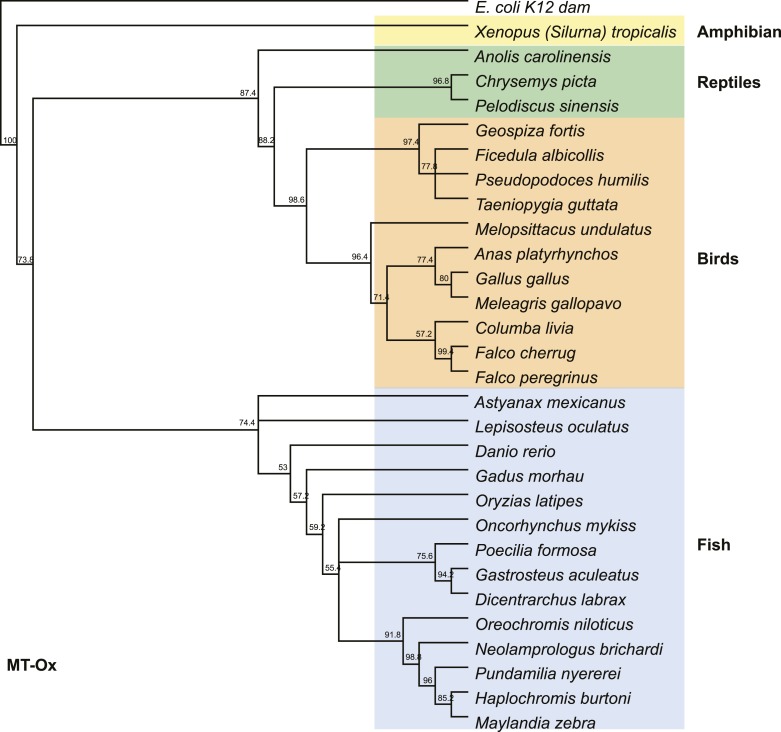
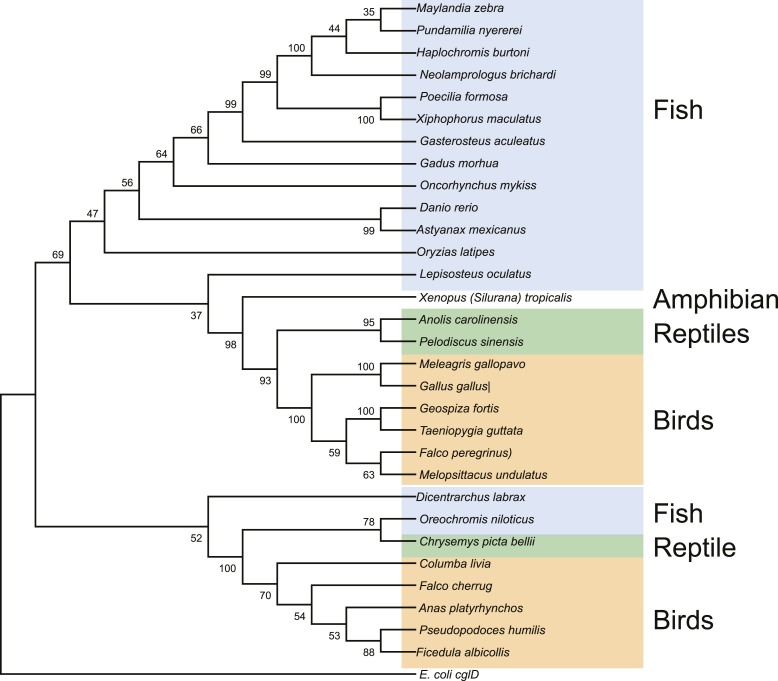
Figure 3. Model timeline for the evolution of the EEVS and MT-Ox genes in vertebrates.
The genes entered an early vertebrate genome as a linked pair (vertical blue arrow) and were retained in the modern ray-finned fish, amphibians, reptiles, and birds as indicated by thick dark cyan arrows. Coelacanths and mammals lost the genes (thick red arrows). No full genome sequence is available for assessing the presence of EEVS and MT-Ox in lungfish. The phylogenetic trees of the EEVS and MT-Ox proteins or mRNA from a selected set of vertebrates can be found in Figure 3—figure supplements 1–3 and Supplementary files 1, 2.