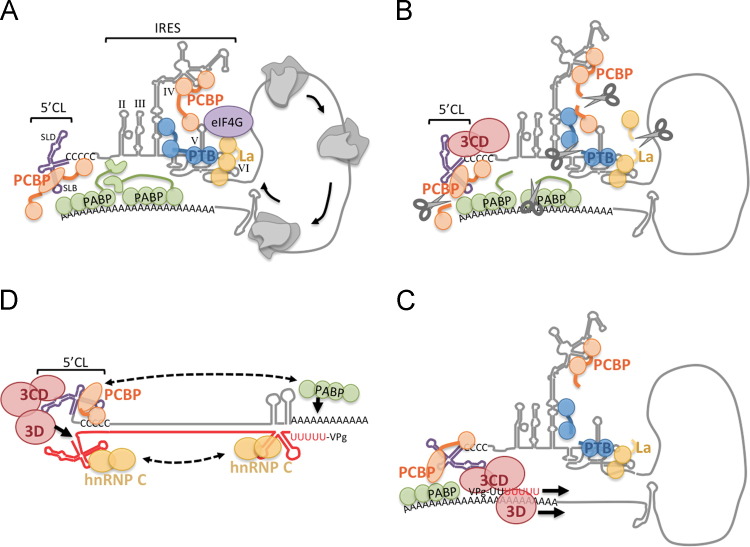
Fig. 1.
Cartoon illustrating roles of RBPs in PV replication. (A) PV Translation. Host factor PCBP binds both cloverleaf stem-loop B (SLB) and IRES SLIV, PTB binds SLV and together with La helps eIF4G and other canonical translation factors (not shown) bind properly to recruit ribosomes. ITAFs provide RNA chaperone functions and modular binding motifs link and fold different stem loops. Interactions of PABP with PCBP and PABP with initiation factors (eIF4G, eIF4B interactions not depicted) help close the RNA 5′-3′ loop and facilitate ribosomes recycling from stop codon to start codon. Other ITAFs for PV mentioned in the text are not depicted. (B) Switch to replication. Viral polymerase precursor 3CD binds 5′CL SLD, together with cleavage of PCBP, PTB and La by 3Cpro (scissors), converts the template to translation–initiation–incompetent state, plus blocks ribosome recycling and thus ribosomes runoff and clear the template. (C) PV minus strand replication. Replicase complex consisting of interacting 3CD–3Dpol complexes initiates replication. The complex is oriented properly via PCBP binding PABP. (D) PV plus strand replication. The double-stranded RF replication intermediate (nascent negative strand shown in red) breathes allowing cloverleaf and anti-cloverleaf to form, stabilized by binding PCBP and hnRNP C respectively. Polymerase replicase complex also builds on 5′ CL and initiates replication on negative strand template that is properly positioned for precise-end initiation. hnRNP C can also interact with SL on the 5′ end of the negative strand that also requires breathing to form. In this scenario PABP may be able to rebind to poly(A) tail and hnRNP oligomerization and PABP interaction with PCBP may facilitate genome circularization in double-stranded RF intermediate.