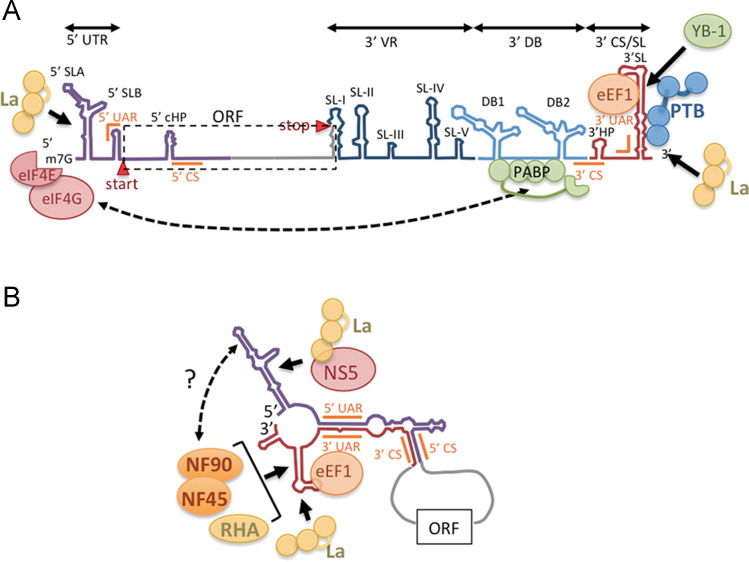
Fig. 2.
(A) Structural elements of flavivirus 5′ and 3′ UTRs based on Dengue virus sequences. Start and stop codons defining the open reading frame are shown. The 5′ UTR is ~95 nt and includes 5′ SLA, 5′ SLB. The flavivirus 3′ UTR is <380 nt and contains regions with conserved structural elements: VR (dark blue) with SL-I through IV, the 3′ DB region containing duplicate dumbbell structures (DB1 and DB2) (light blue) and the 3′ CS/SL region (red) containing the conserved 3′ SL. All structures from SL-1 through DB-2 participate in pseudoknots (not depicted). To facilitate translation poly(A)-binding protein binds A-rich sequences in the DB region allowing protein-bridge looping to translation factor eIF4G associated with the 5′ cap structure. YB-1 binds the 3′ SL and represses translation and replication. PTB and La bind 3′ UTR may also facilitate translation. (B) Long range interaction between complimentary sequences in 5′ and 3′ regions (5′ CS, 3′ CS; 5′ UAR, 3′ UAR) facilitate genomic looping associated with RNA replication. This looping remodels RNA into new conformations that may promote binding of additional host RBPs not associated with translating DENV RNA such as eEF1, and the dsRNA binding proteins NF90, NF45 and RHA that bind the 3′ SL. In HCV, NF90 binds both 5′ and 3′ UTR of HCV RNA and RHA is involved in bridging 5′ and 3′ UTRs (not depicted). A similar arrangement may exist in flaviviruses where NF90 and RHA may also bind 5′ UTR stem loops and stabilize the looped structure to promote negative strand RNA replication. The NS5 RdRp binds to the 5′ SL1 which due to looping helps position the polymerase near the 3′ terminus (Bidet and Garcia-Blanco, 2014). NS5 may also be recruited by La, which binds the 5′ UTR.