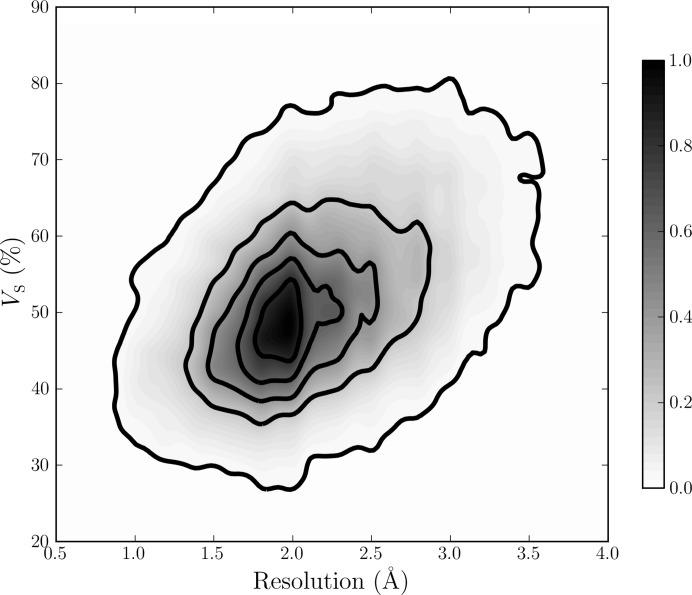
Figure 2.
Two-dimensional density function of V S for 60 218 protein crystal forms using binned two-dimensional kernel estimates. The scale of V S from 20 to 90% on the y axis corresponds to V M values between 1.54 and 12.3 Å3 Da−1. The plot has been normalized to have a maximum value of 1. Isocontour lines are drawn as solid bold lines in increments of 0.2. There is a clear trend towards lower values of V S at higher resolutions. Most density is centred about approximately 1.9 Å resolution and V S = 50%, and typical values for V S range between 30 and 80%. Figure from Weichenberger & Rupp (2014 ▶). Owing to a 60-fold lower number of available DNA/RNA crystal structures in the PDB, the dependence of experimental resolution on solvent content cannot be established for nucleic acid chains (see Supporting Information).