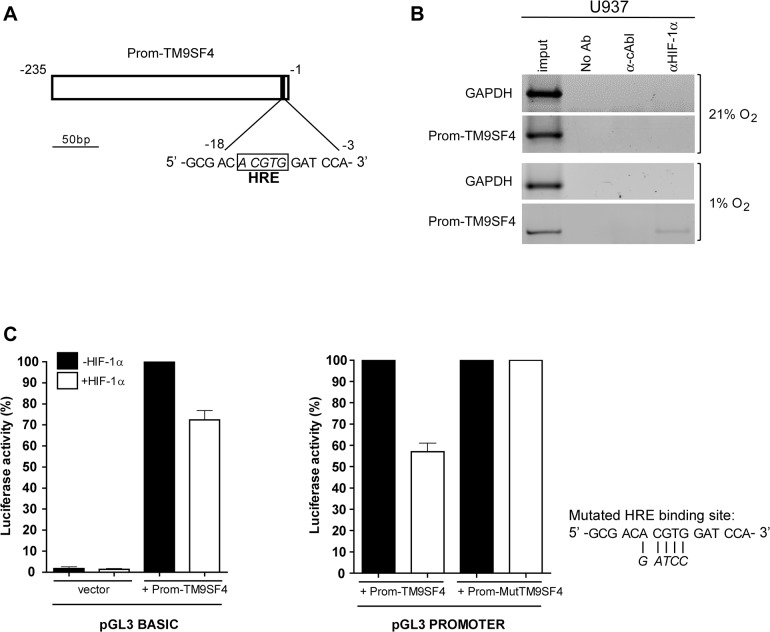
Fig 5. In hypoxia, TM9SF4 is a direct target gene of HIF-1α in leukemic cells.
(A) HIF-1α DNA binding site (HRE) identified on TM9SF4 partial promoter sequence is shown. (B) Chromatin immunoprecipitation (ChIP) experiments were performed by using protein extracts of U937 cells cultured in hypoxia (panels 1% O2), as compared to normoxia (panels 21% O2), immunoprecipitated with the anti(α)-HIF-1α or with an unrelated α-cAbl antibodies, or no antibody (No Ab), and analyzed by PCR amplification of the TM9SF4 region surrounding the HRE site (Prom-TM9SF4) or a GAPDH coding region used as an internal control. Input: PCR on 0.02% of total chromatin. One representative experiment of three is shown. (C) Promoter activity assays were performed first (left panels) by using TM9SF4 upstream DNA sequence subcloned in the pGL3Basic promoterless luciferase vector (Prom-TM9SF4) and transfected into 293T cells in the presence (white bars) or absence (black) of a HIF-1α expression vector, as compared to empty pGL3Basic vector (vector); then (right panels) by using Prom-TM9SF4, or the same region with a mutated HRE binding site (Prom- MutTM9SF4), cloned in the pGL3Promoter reporter vector containing a minimal promoter, and transfected with or without a HIF-1α expression vector. Data of luciferase activity detected are mean ± S.E.M. values of 3 independents experiments.