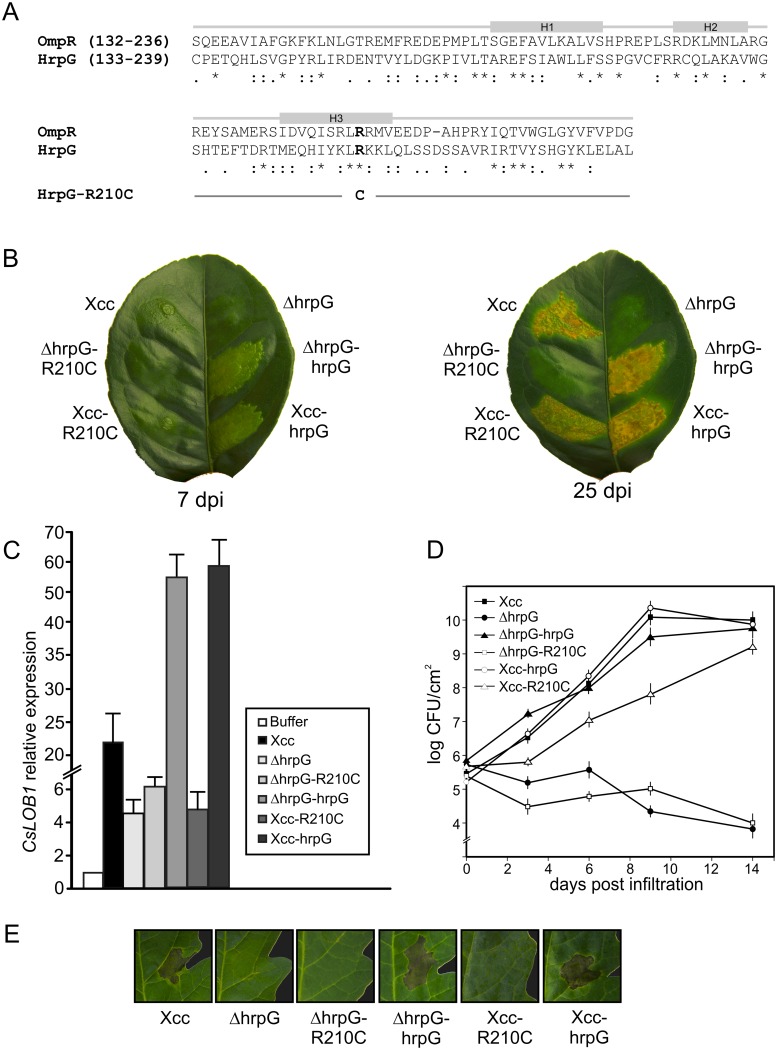
Fig 1. Characterization of the residue Arg210 of HrpG in Xcc pathogenicity and HR induction.
(A) Sequence alignment and secondary structure assignments of a region of the DNA-binding domain of OmpR from E. coli and HrpG from Xcc. Helices α1, α2 and α3 are depicted with grey boxes. Asterisks (*) indicate identical residues, colons (:) are conservative replacements and full stops (.) are semiconservative replacements. Arg209 in OmpR sequence and Arg210 in Xcc sequence are depicted in bold. This residue turns into Cys210 in HrpG-R210C sequence (B) Xcc wild type, deletion mutant ΔhrpG, complemented strains ΔhrpG-R210C and ΔhrpG-HrpG and Xcc carrying the wild type and R210C mutant copy, Xcc-HrpG and Xcc-R210C, respectively, were inoculated at 107 CFU/ml in 10 mM MgCl2 into the intercellular spaces of fully expanded citrus leaves. Representative leaves are shown 7 dpi (left panel) and 25 dpi (right panel). (C) RT-qPCR to determine CsLOB1 expression levels in leaves after 24 hours of inoculation with Xcc strains. Bars indicate the expression levels relative to buffer infiltrations. Values are the means of four biological replicates with three technical replicates each. (D) Bacterial growth of the Xcc strains in citrus leaves. Values represent the mean of three samples from three different plants. Error bars indicate standard deviations. (E) Xcc variants were inoculated at 107 CFU/ml in 10 mM MgCl2 in tomato leaves and a representative photograph after 24 h is shown.