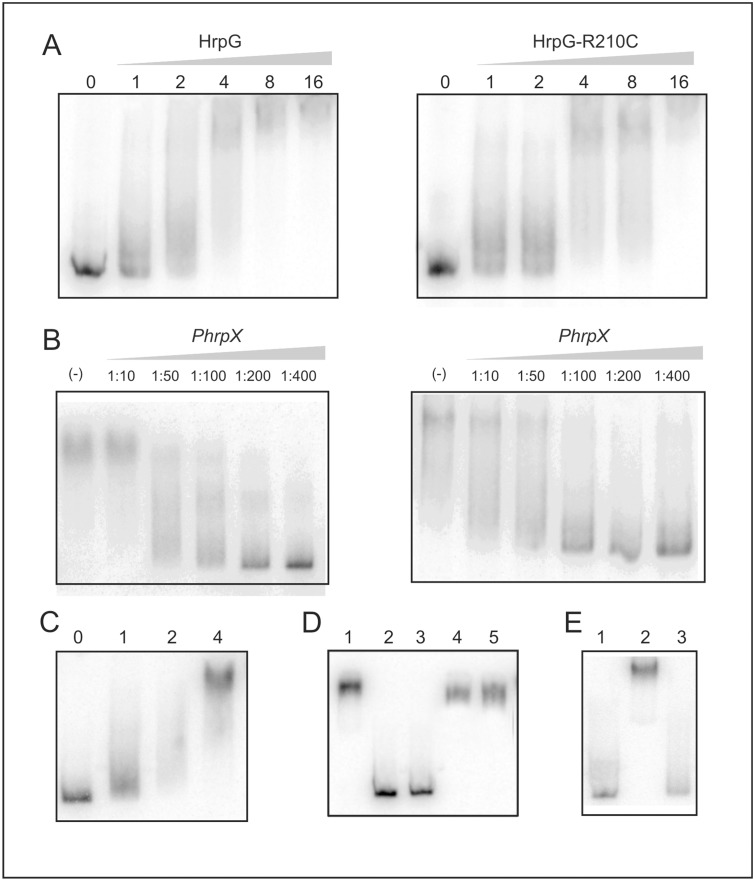
Fig 3. A substitution of Arg210 by cysteine does not prevent binding to DNA promoter regions.
(A) Electrophoretic mobility shift assay of 32P-labeled hrpX promoter and purified HrpG and HrpG-R210C. Numbers in the top of the lanes indicate pmoles of protein added to the assay. (B) Complex competition assays with excess unlabeled DNA: Lanes labeled (-) show the complex between HrpG (left panel) and HrpG-R210C (right panel) with 8 pmol of protein. Competition was performed using the ratio indicated in each lane. (C) EMSA of 32P-labeled hrpX promoter and a mixture 1:1 of HrpG and HrpG-R210C with the total pmol indicated on top of each lane. (D) Control EMSA of 32P-labeled hrpX promoter and purified Trx in the same conditions as HrpG to discard unspecific interactions. Lane 1 shows the binding of 16 pmoles of HrpG to PhrpX as control. Lanes 2 and 3: PhrpX was incubated with 15 and 30 pmoles of Trx, respectively in the same conditions as for HrpG. Lanes 4 and 5: unspecific competition assay in which the HrpG-PhrpX complex was challenged with 200 ng of poly-dIdC (lane 4) or salmon sperm DNA (lane 5). (E) Lane 1: free probe, lane 2: binding of 16 pmoles of HrpG to PhrpX as control, lane 3: PhrpX was incubated with 25 μg of a protein extract obtained from E. coli bearing the pET32 empty vector.