Figure 1.
RIG-I and MDA5 can displace viral proteins bound to dsRNA in an ATP-dependent manner.
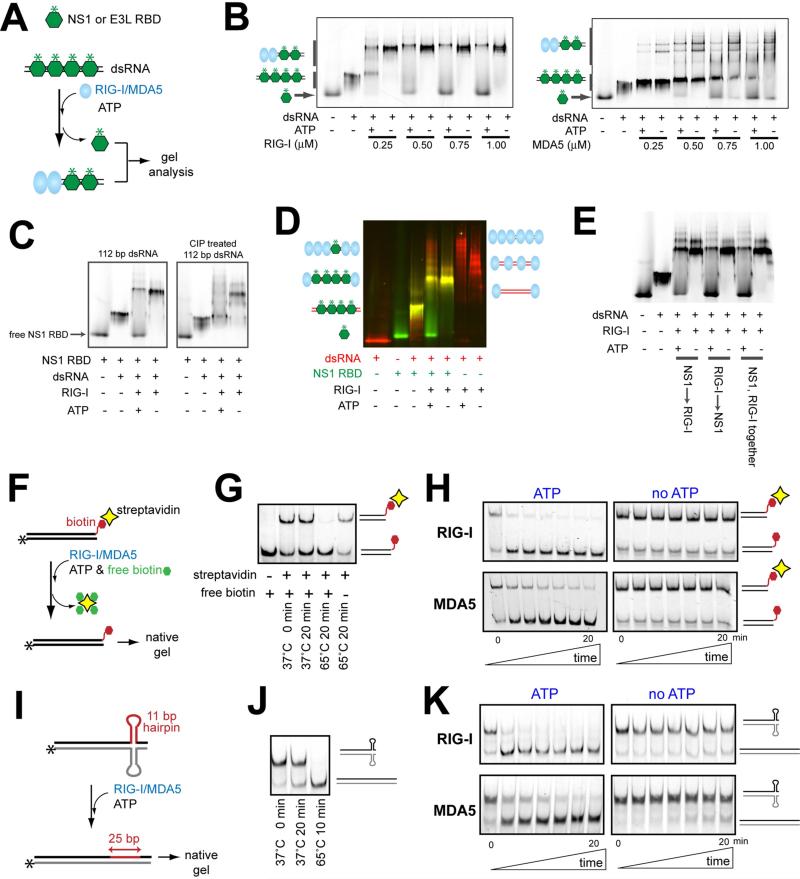
A. Schematic of the viral protein displacement assay. The in vitro transcribed 112 bp dsRNA (45 nM) was pre-incubated with a saturating concentration (1 μM) of fluorescently labeled (*) RBDs of NS1 and E3L, and further incubated with RIG-I or MDA5 in the presence or absence of ATP. Unless mentioned otherwise, all dsRNAs in this manuscript contain 5’ppp at both ends.
B. Gel analysis of the protein displacement assay using NS1 RBD as described in (A). Gel images were obtained using fluorescein conjugated to NS1 RBD. For results with E3L RBD, see Figure S1B.
C. The NS1 displacement assay using RIG-I and 112 bp dsRNA before and after CIP treatment.
D. The NS1 displacement assay using RIG-I and 3’-Cy3-labeled 112 bp dsRNA (before CIP). The gel image was obtained by overlaying the images obtained using the Cy3 fluorescence (red) and the fluorescein fluorescence (green).
E. The NS1 displacement assay using RIG-I and 112 bp dsRNA. NS1 RBD and RIG-I were incubated with dsRNA in the indicated order.
F. Schematic of the streptavidin displacement assay. The 3’-biotinylated 112 bp dsRNA was complexed with streptavidin and then with RIG-I/MDA5. The displacement reaction was initiated with ATP and an excess amount of free biotin (streptavidin trap) at 37°C, and quenched with EDTA prior to gel analysis.
G. Stability of the streptavidin:biotinylated dsRNA interaction at 37°C.
H. Gel analysis of the streptavidin displacement as described in (F), using the fluorescence of RNA.
I. Schematic of RNA hairpin displacement assay. The 112 bp dsRNA containing the complementary hairpins (Figure S1F) was complexed with RIG-I/MDA5 in the presence or absence of ATP at 37°C prior to gel analysis.
J. Stability of the hairpin containing duplex at 37°C.
K. Gel analysis of the hairpin displacement as described in (I), using the fluorescence of RNA.