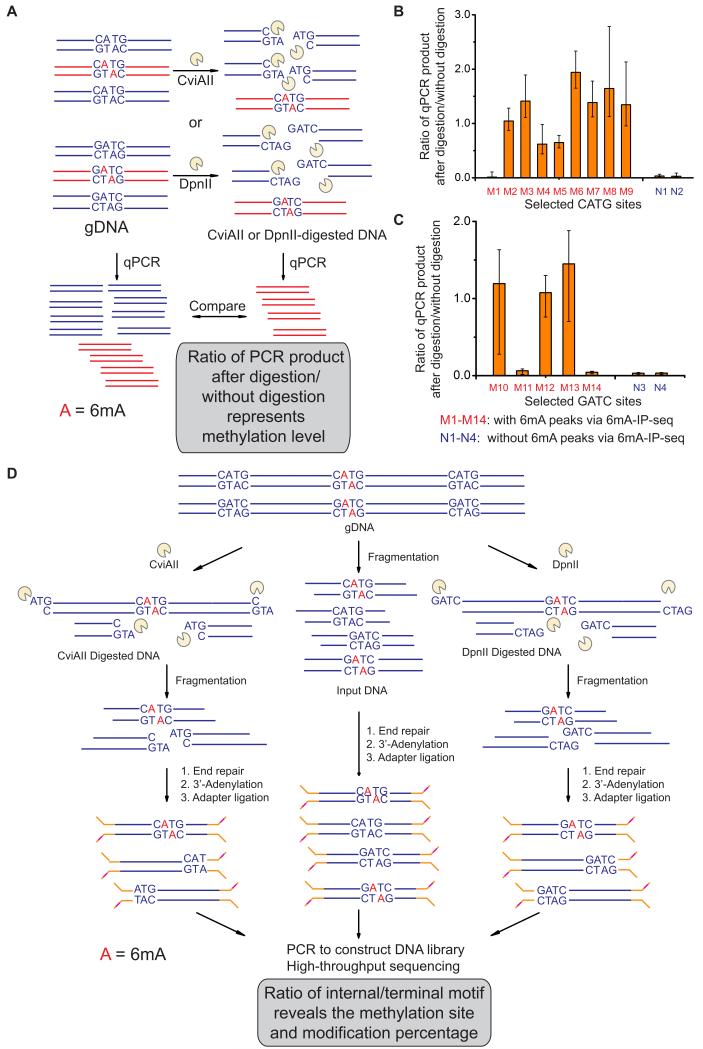
Figure 4. Single Site Detection of 6mA Using Methylation Sensitive Restriction Enzymes.
(A) Schematic diagram of 6mA-RE-qPCR for validation of specific 6mA. Restriction enzymes CviAII or DpnII that are sensitive to 6mA methylation in CATG or GATC were used to digest the unmethylated CATG or GATC sites in genomic DNA, respectively. The undigested CATG or GATC sites represent the methylated fraction, and can be PCR-amplified by using primers that cover these sites.
(B, C) qPCR results of 11 selected CATG sites and 7 GATC sites validated the accuracy of 6mA-IP-seq. After CviAII- or DpnII-mediated digestion, qPCR was performed using specific primers covering these sites. Relative abundances of undigested CATG or GATC sites were calculated from the ΔCt value between digested and undigested DNA samples (n = 3, mean ± S.E.M.).
(D) Schematic diagram of 6mA-RE-seq. gDNA is digested with CviAII or DpnII, sonicated to small fragments around 100 base pair, and constructed into sequencing libraries. The ratio for CATG or GATC internal of sequence reads versus at the end of sequence reads of a specific genomic site represents the relative methylation to unmethylation ratio. An input sample from gDNA without CviAII- or DpnII-based digestion serves as a control.