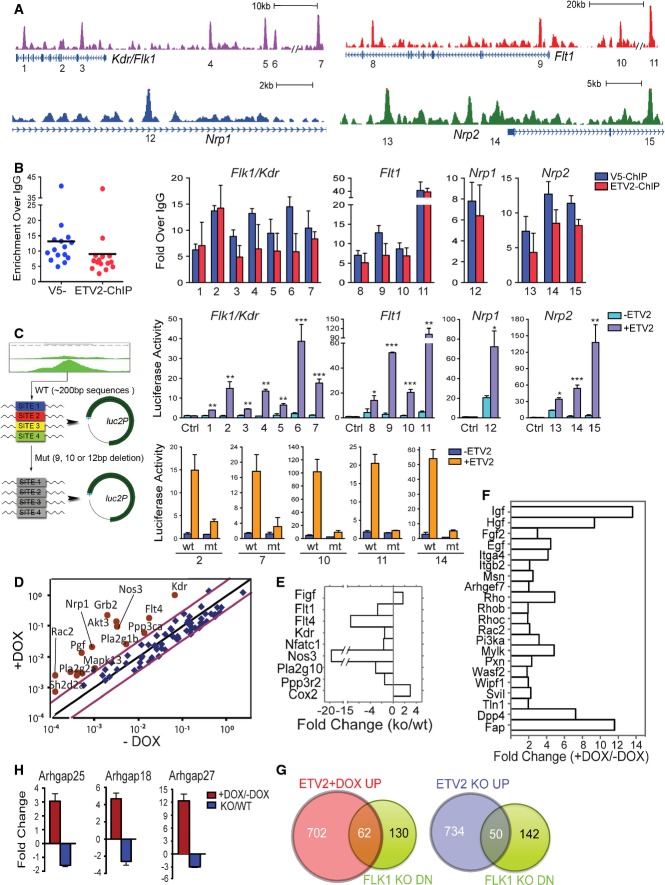
Genomic snapshots depicting the ETV2 binding peaks associated with the VEGFR genes. Numbers indicate ChIP-Seq peaks with high confidence.
ChIP-PCR analysis showing ETV2 recruitment to the 15 potential binding sites in (A). ETV2 enrichments were shown by both V5 antibody and ETV2-polyAb pull-down. PCR primers and genomic locations are provided in Supplementary Table S3. Error bars represent SD, n = 4.
Luciferase reporter assay for 1–15 ETV2 peak regions from (A). The indicated peak regions or selected ETV2 binding motif deletion mutants (2, 7, 10, 11, and 14) were cloned into the pGL4.24[luc2P/minP] vector (left panel). 293T cells were transfected with pGL4.24 control vector, ETV2 wild-type, or mutant luciferase reporter constructs, together with Renilla luciferase vector in the presence (pMSCV-Etv2) or absence of ETV2 expression plasmids. For each ETV2 peak reporter construct, luciferase activity was first normalized to Renilla luciferase values. Luciferase activity value obtained with ETV2 was then compared to that of the –ETV2 value. Error bars represent the SEM obtained from four biological replicates. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed Student's t-test.
Mouse VEGF signaling array using RNA from day 3.5 iEtv2 EBs (±DOX from day 2) is shown as scattered plots of fold changes (+DOX versus −DOX). Each gene was normalized to three housekeeping genes. Red dots are upregulated genes by DOX addition.
RNA obtained from E8.5 Etv2+/+ and Etv2−/− embryos was subjected to VEGF signaling analysis. Key genes with significant changes in expression are shown.
Mouse Cell Motility PCR array using RNA from day 3.5 iEtv2 EBs (±DOX from day 2) is shown as fold changes (+DOX versus −DOX).
Venn diagram showing an overlap between downregulated genes in E8.5
Flk1−/− embryos
41 and upregulated genes by ETV2 overexpression (left) or ETV2 knockout (right)
27.
Gene expression analysis of Rho-GTPase activating proteins 18, 25, and 27 in day 3 iEtv2 (±DOX on day 2) or Etv2+/+ and Etv2−/− EB cells. Genes were normalized to Gapdh. The fold change was obtained from the ratios of +DOX to −DOX or Etv2−/− to Etv2+/+. Error bars represent the SD of four independent biological samples.