Abstract
Sigma54 (σ54) regulates nitrogen and carbon utilization in bacteria. Promoters that are σ54-dependent are highly conserved and contain short sequences located at the −24 and −12 positions upstream of the transcription initiation site. σ54 requires regulatory proteins known as bacterial enhancer-binding proteins (bEBPs) to activate gene transcription. We show that σ54 regulates the capacity to grow on various nitrogen sources using a Bacillus thuringiensis HD73 mutant lacking the sigL gene encoding σ54 (ΔsigL). A 2-fold-change cutoff and a false discovery rate cutoff of P < 0.05 were used to analyze the DNA microarray data, which revealed 255 genes that were downregulated and 121 that were upregulated in the ΔsigL mutant relative to the wild-type HD73 strain. The σ54 regulon (stationary phase) was characterized by DNA microarray, bioinformatics, and functional assay; 16 operons containing 47 genes were identified whose promoter regions contain the conserved −12/−24 element and whose transcriptional activities were abolished or reduced in the ΔsigL mutant. Eight σ54-dependent transcriptional bEBPs were found in the Bt HD73 genome, and they regulated nine σ54-dependent promoters. The metabolic pathways activated by σ54 in this process have yet to be identified in Bacillus thuringiensis; nonetheless, the present analysis of the σ54 regulon provides a better understanding of the physiological roles of σ factors in bacteria.
Keywords: Bacillus thuringiensis, sigma54, bEBPs, metabolic pathways, DNA microarray
Introduction
Sigma (σ) factors are a class of proteins constituting dissociable subunits of prokaryotic RNA polymerase. Different σ factors associate with the core polymerase and recognize the promoters of target genes to initiate transcription in response to environmental conditions. Identifying regulons in bacteria that are controlled by specific σ factors is important for understanding cell responses to given stimuli.
The sigL gene (known as rpoN gene in Gram-negative soil bacteria) encodes σ54. Promoters that are σ54-dependent are highly conserved and contain short sequences located at the −24 and −12 positions upstream of the transcription initiation site, in contrast to σ70-dependent promoters containing upstream sequences located at −35 and −10 (Buck et al., 2000). As observed for eukaryotic enhancer-binding proteins (EBPs), σ54 requires regulatory proteins known as bacterial (b)EBPs to activate gene transcription (Bush and Dixon, 2012).
σ54 activates genes that are involved in the utilization of nitrogen and carbon for energy and a range of other cellular processes in bacteria (Merrick, 1993). The Sigma 54 regulon is found in several species, including Escherichia coli (Reitzer and Schneider, 2001), Pseudomonas putida (Cases et al., 2003), Vibrio cholera (Dong and Mekalanos, 2012), Xylella fastidiosa (da Silva Neto et al., 2010), Geobacter sulfurreducens (Leang et al., 2009), Listeria monocytogenes (Arous et al., 2004), and several members of Rhizobiaceae family (Dombrecht et al., 2002; Hauser et al., 2007). E. coli has approximately 30 σ54-dependent operons, about half of which are involved in nitrogen assimilation and metabolism; the others participate in the conservation of metabolites and energy resources under adverse conditions (Reitzer and Schneider, 2001), and are related to arginine and histidine transport (Caldara et al., 2007), arginine catabolism (Kiupakis and Reitzer, 2002), acetoacetate catabolism (Matta et al., 2007), nitrogen assimilation (Magasanik, 1989; Yamada et al., 2007), glutamine transport (Baev et al., 2006), and propionate catabolism (Lee et al., 2005). In V. cholera, 68 σ54-binding sites and 82 operons positively regulated by σ54 have been identified, of which 37 binding sites are confirmed by chromatin immunoprecipitation (Dong and Mekalanos, 2012).
Among Gram-positive bacteria, 77 genes have been identified in L. monocytogenes, most of which are related to carbohydrate (e.g., pyruvate) metabolism (Arous et al., 2004). Several metabolism-related operons regulated by σ54 have also been identified in Bacillus subtilis such as those involved in isoleucine and valine utilization (Debarbouille et al., 1999), the acetoin (3-hydroxy 2-butanone) catabolic pathway (Ali et al., 2001), and arginine catabolism (Heidrich et al., 2006), as well as in the degradation of polymers of fructose (levanes) (Debarbouille et al., 1991a).
B. anthracis, B. cereus, and B. thuringiensis (Bt) are all spore-forming members of the B. cereus group (Helgason et al., 2000). These species vary in terms of host range and virulence (Han et al., 2006), and are mainly distinguished by the genes contained in their plasmids. Bt forms parasporal crystals during the stationary phase of growth that are toxic to a wide variety of insect larvae (Schnepf et al., 1998), making Bt strains the most commonly used biological pesticide worldwide. Among these species, only two metabolic pathways are known to be controlled by σ54: the γ-aminobutyric acid (GABA) (Zhu et al., 2010) and l-lysine metabolism (Zhang et al., 2014) pathways in the Bt HD73 strain. Little is known about the roles of σ54 and its regulons in the B. cereus group. Thus, the aim of the present study was to investigate these roles using the HD73 strain of Bt.
Regulons and binding sites of σ54 were identified by DNA microarray and computational predictions, which were also used to analyze EBP domains and gene organization and predict operons. The results revealed several novel σ54-dependent metabolic pathways. These findings provide insight into σ54-dependent regulation in the B. cereus group and demonstrate that the σ54 regulon in Bt differs from those of other Gram-positive bacteria.
Materials and methods
Bacterial strains, plasmids, and culture
Bacterial strains and plasmids used in this study are listed in Additional file 4. E. coli TG1 was used for cloning, while ET12567 was used to produce unmethylated plasmid DNA for Bt transformation (Wang et al., 2006). Wild-type Bt HD73 (laboratory stock) expressing cry1Ac was used throughout this study. Bt strains were transformed by electroporation as previously described (Wang et al., 2006). E. coli was grown at 37°C in Luria-Bertani (LB) medium (1% NaCl, 1% tryptone, and 0.5% yeast extract). Bt was grown in LB medium, Schaeffer's sporulation medium (SSM) (Schaeffer et al., 1965), or glucose minimal medium (GMM) (Debarbouille et al., 1991b) supplemented with 40 mM of a given amino acid as the sole nitrogen source, with vigorous shaking (220 rpm) at 30°C. The antibiotic concentrations used for bacterial selection were as follows: 100 μg/ml ampicillin for E. coli, 5 μg/ml erythromycin and 50 μg/ml kanamycin for Bt. Bacteria producing β-galactosidase were identified by culturing in SSM medium.
DNA manipulation techniques
Plasmid DNA was extracted from E. coli by a standard alkaline lysis procedure with a plasmid miniprep kit (Axygen Scientific, Union City, CA, USA). Chromosomal DNA was extracted from Bt cells as previously described (Lereclus et al., 1989). Restriction enzymes and T4 DNA ligase were used according to the manufacturer's instructions (New England Biolabs, Ipswich, MA, USA). Oligonucleotide primers were synthesized by Sangon (Shanghai, China); all primer sequences are listed in Additional file 5. PCR was performed with high-fidelity DNA polymerase (Toyobo, Osaka, Japan). Amplified fragments were purified with a PCR cleanup kit (Axygen). Digested DNA fragments were separated on 1% agarose gels and extracted using a DNA gel extraction kit (Axygen). All constructs were confirmed by sequencing (Invitrogen, Carlsbad, CA, USA).
Preparation of RNA for DNA microarray
Total RNA was extracted from Bt cells grown in SSM medium at stage T7 (7 h after the end of the exponential phase). Three independent repeats from different clones were performed for each strain. Cells were harvested from 1 ml of culture by centrifugation (13,000 × g for 30 s at 4°C), and cell pellets were immediately suspended in 1 ml cold TRI-Reagent (Invitrogen). The suspensions were then snap-frozen in liquid nitrogen. The RNA was extracted using the Qiagen Easy RNA kit (Hilden, Germany). Residual DNA was removed using RNase-free DNase I (New England Biolabs), and the resulting RNA samples were stored at −80°C.
RNA amplification, labeling, and hybridization
Total RNA was amplified and labeled with the Low Input Quick Amp Labeling Kit, One-Color (Agilent Technologies, Santa Clara, CA, USA). Labeled RNA was purified using the RNeasy mini kit (Qiagen). Each slide was hybridized with 1.65 μg Cy3-labeled cRNA using the Gene Expression Hybridization Kit in a hybridization oven (both from Agilent Technologies). After 17 h, the slides were washed in staining dishes (Thermo Fisher Scientific, Waltham, MA, USA) using the Gene Expression Wash Buffer kit (Agilent Technologies).
Data acquisition
Slides were scanned using a Microarray Scanner (Agilent Technologies) with default settings (dye channel: green; scan resolution: 5 μm; photomultiplier tube: 100%; 10%, 16-bit) and Feature Extraction software 10.7 (Agilent Technologies). Raw data were normalized with the quantile algorithm of Gene Spring Software 11.0 (Agilent Technologies). Transcriptome data were analyzed with statistical tests.
Construction of −12/−24 promoter with lacZ fusions
The predicted −12/−24 regions of promoters were amplified from Bt HD73 genomic DNA using specific primers. Promoter restriction fragments were then ligated into the pHT304-18Z vector containing a promoterless lacZ gene (Agaisse and Lereclus, 1994). Recombinant pHT-Pn (where n indicates the gene ID in the Bt HD73 genome) was introduced into Bt HD73 and ΔsigL mutant strains. The resultant strains, HD73(Pn) and ΔsigL(Pn), were selected by resistance to erythromycin and tested by PCR to confirm the presence of the promoter fragments in the plasmids.
β-galactosidase assay
Bt strains were cultured in SSM medium at 30°C and 220 rpm. A 2-ml volume was collected at 1-h intervals from T1 to T8, from which cells were harvested by centrifugation for 1 min at 10,000 × g. The supernatant was removed, and the pellet was stored at −20°C or resuspended in 500 μl Buffer Z with 1 mM dithiothreitol. The β-galactosidase activity was determined as previously described (Perchat et al., 2011) and expressed as Miller units. Reported values represent averages from at least three independent assays.
Construction of EBP mutants
Five EBP (rocR, prdR, acoR, bkdR, and levR) deletion strains were generated by allelic exchange as previously described (Zhu et al., 2010) using a kanamycin resistance cassette (kan, 1473 bp) and the thermosensitive suicide plasmid pMAD (Arnaud et al., 2004). The upstream and downstream regions (fragments A and B, respectively) of the hd_0559 (rocR) gene were amplified by PCR using chromosomal DNA from Bt as template and rocR-AF/rocR-AR and rocR-BF/rocR-BR primers. The kan gene was amplified by PCR using pDG780 (Guerout-Fleury et al., 1995) as template and rocR-kmF/rocR-kmR primers. Fragment A and kan were ligated together by overlapping PCR using primers rocR-AF and rocR-kmR. The amplification product was then integrated with fragment B in a second round of overlapping PCR using primers rocR-AF and rocR-BR. The resultant PCR products were digested, purified, and ligated with the pMAD plasmid to generate pMADΔrocR. The host strain was transformed with the recombinant plasmid by electroporation, and erythromycin-sensitive transformants were selected. The correct mutant strain was identified by PCR. The hd_1069 (prdR), hd_3228 (acoR), hd_4469 (bkdR), and hd_5607 (levR) deletion strains were constructed as described above.
The hd_3141 (soxR) deletion mutant was constructed as follows. DNA fragments corresponding to upstream and downstream regions of the soxR gene were amplified by PCR using genomic DNA from Bt HD73 as the template and the soxR-a/soxR-d and soxR-b/soxR-c primers. The amplified fragments were fused via overlapping PCR using the soxR-a/soxR-b primers. The resultant 1257-bp fragment was then digested with the BamHI and EcoRI restriction enzymes and inserted between the corresponding restriction sites of the pMAD plasmid. The recombinant pMADΔsoxR plasmid was electroporated into host strains and erythromycin-sensitive transformants were selected.
Results
Characterization of the sigL mutant of Bt
Individual l-amino acids were tested for their ability to support the growth of HD73 and ΔsigL mutant strains in a GMM. These two strains were unable to use alanine, aspartate, asparagine, glutamic acid, glutamine, threonine, cysteine, phenylalanine, tyrosine, glycine, lysine, and methionine as nitrogen sources (data not shown), but were able to use histidine and arginine (Table 1). In addition, the ΔsigL mutant was unable to use sarcosine, leucine, isoleucine, serine, or valine as nitrogen sources, unlike the HD73 strain (Table 1). These results suggest that σ54 controls the pathways responsible for the utilization of these amino acids.
Table 1.
Doubling time of HD73 wild type and ΔsigL mutant strains grown in minimal medium containing various nitrogen sources.
| Nitrogen source | Doubling time (hour) | |
|---|---|---|
| HD73 | ΔsigL | |
| Sarcosine | 16.54 | >60 |
| Proline | 30.13 | >60 |
| Histidine | 15.93 | 16.91 |
| Leucine | 18.65 | >60 |
| Isoleucine | 10.69 | >60 |
| Serine | 16.84 | >60 |
| Arginine | 9.71 | 8.75 |
| Valine | 14.74 | >60 |
Functional classification of differentially expressed genes in wild-type and ΔsigL mutant strains
To further investigate the function of σ54 in Bt, the transcriptome of HD73 and ΔsigL mutant strains was compared in order to identify potential transcriptional targets of σ54. A 2-fold-change cutoff and a false discovery rate cutoff of P < 0.05 were used to analyze the DNA microarray data, which revealed 255 genes that were downregulated and 121 that were upregulated in the ΔsigL mutant relative to the wild-type HD73 strain. DNA microarray data were deposited at the NCBI Gene Expression Omnibus (accession no. GSE48410). A complete list of the differentially expressed genes and their expression ratios is provided as Additional file 1. The genes were assigned to five functional groups according to Kyoto Encyclopedia of Genes and Genomes, Clusters of Orthologous Groups, and Pfam protein family classifications of the Bt subsp. kurstaki strain HD73 genome, most encoding hypothetical proteins or those of unknown function proteins (117 and 72 that were down- and upregulated, respectively). The top three categories comprised genes associated with amino acid metabolism (66 down- and five upregulated), carbohydrate metabolism (17 down- and 15 upregulated) and intracellular transport system (29 down- and 11 upregulated). Other genes were associated with sporulation (17 down- and two upregulated) and signal transduction (nine down- and 16 upregulated). More than 70% of the genes were associated with amino acid and carbohydrate metabolism.
Validation of predicted σ54-related promoter elements with transcriptional lacZ fusion constructs
A computational analysis was carried out to identify σ54 recognition sequences within the HD73 genome. The conserved sequence BYGGCMYRNNNNYYGCW (Francke et al., 2011) was searched 700 bp upstream and 100 bp downstream of start codons. After eliminating the coding regions, 14 putative σ54-binding sites were found in the same strand orientation as their potential target genes (Additional file 2). Of these, 10 were located in genes that were downregulated in the DNA microarray data and one was in an upregulated gene, which was fewer than expected. To identify genes potentially controlled by σ54 from an expanded pool, a search for the minimally conserved sequence NNGGN10GCNN (critical bases underlined) was carried out in the region 700 bp upstream of the start codon of genes that were downregulated in the DNA microarray. Using this approach, 17 promoters were identified containing the target sequence, including the 10 putative σ54-binding sites previously predicted with the highly conserved sequence, for a total of 21 genes potentially regulated by σ54 (Additional file 2).
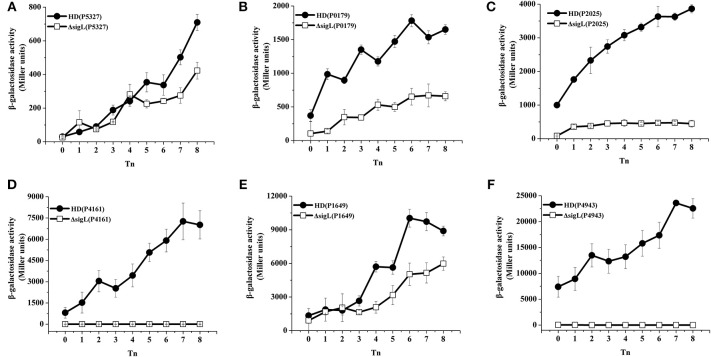
To validate the results of the transcriptome and −12/−24 region analyses, 21 promoter fusions with the lacZ gene were constructed and analyzed by evaluating β-galactosidase activity. The activation of 16 promoters was abolished and decreased in the ΔsigL mutants containing the gabT (Zhu et al., 2010) and kamA (Zhang et al., 2014) promoters, these promoters are regulated by σ54 (Figures 1A–F, 2A–H). Three of these (HD73_3213, HD73_4468, and HD73_5614) were regulated by sequences that were highly similar to B. subtilis SigL promoter elements (Debarbouille et al., 1991a, 1999; Ali et al., 2001). One promoter (P2953) containing the highly conserved −12/−24 region was expressed neither in HD73 nor in the ΔsigL mutant. The activation of two of the promoters (P2699 and P4960) was unaffected and two (P0035 and P1772) showed increased expression in the ΔsigL mutant, suggesting that they are not directly controlled by σ54 (data not shown). Finally, the σ54 regulon of the Bt HD73 strain included five genes and 11 operons containing a total of 47 genes (Table 2, the annotation of these genes is described in Table 3); of these, 12 had highly conserved −12/−24 promoter sequences and four had minimally conserved sequence. The 16 promoters sequences were aligned and the conserved sequence WYGGHDYRNHDNNWGCD was identified for σ54-regulated genes.
Figure 1.
β-galactosidase activity assay of the putative −12/−24 promoters. Wild-type strain HD73 (•) and the sigL mutant (□). T0 is the end of exponential phase, and Tn is n hours after T0. (A) Promoter of HD73_5327 with lacZ fusion. (B) Promoter of HD73_0179 with lacZ fusion. (C) Promoter of HD73_2025 with lacZ fusion. (D) Promoter of HD73_4161 with lacZ fusion. (E) Promoter of HD73_1649 with lacZ fusion. (F) Promoter of HD73_4943 with lacZ fusion.
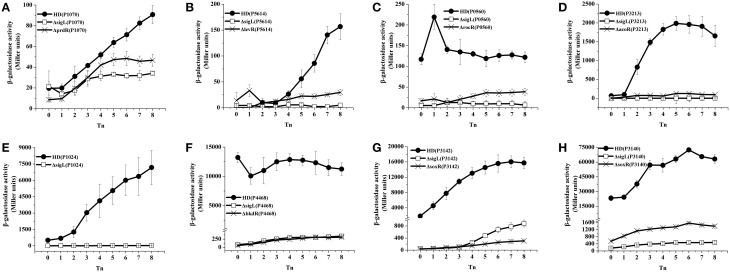
Figure 2.
Identification of EBP target genes. β-galactosidase activity assay of the −12/−24 promoters in wild-type strain HD73 (•), the sigL mutant (□), and the EBP mutants (×). T0 is the end of exponential phase, and Tn is n hours after T0. (A) Promoter of HD73_1070 with lacZ fusion. (B) Promoter of HD73_5614 with lacZ fusion. (C) Promoter of HD73_0560 with lacZ fusion. (D) Promoter of HD73_3213 with lacZ fusion. (E) Promoter of HD73_1024 with lacZ fusion. (F) Promoter of HD73_4468 with lacZ fusion. (G) Promoter of HD73_3142 with lacZ fusion. (H) Promoter of HD73_3140 with lacZ fusion.
Table 2.
Genes or operons controlled by σ54 in Bt.
| Genes or operons | Functions | Promoter −12/−24 Sequence | Position* | EBPs |
|---|---|---|---|---|
| HD73_0366-0369 | γ-aminobutyric acid pathway | TTGGCATACATTTTGCA | −55 | GabR |
| HD73_0560-0562 | Arginine degradative pathway | TTGGTACGTATTTTGCA | −34 | RocR |
| HD73_1024-1025 | Arginine and proline metabolism | TTGGCATGATATTTGCA | −37 | unknown |
| HD73_1070 | Glutamine amidotransferase, class I | TTGGCACGATATTTGCT | −152 | PrdR |
| HD73_2540-2541 | L-lysine metabolic pathway | TTGGCATAACTATTGCT | −38 | KamR |
| HD73_3140-3138 | Sarcosine metabolic pathway | TTGGCATGATTTTTGCA | −41 | SoxR |
| HD73_3142-3147 | Sarcosine metabolic pathway | TTGGCACGTCAATTGCA | −41 | SoxR |
| HD73_3213-3217 | Acetoin degradative pathway | TTGGCACGGTACTTGCA | −37 | AcoR |
| HD73_4161 | Proline metabolism | TTGGCACGCTATTTGCT | −32 | Unknown |
| HD73_4468-4462 | Isoleucine and valine degradation pathway | TTGGCACGGTATTTGCT | −44 | BkdR |
| HD73_5327 | Ubiquinone and terpenoid-quinone biosynthesis | TTGGCATATATGCTGCA | −613 | Unknown |
| HD73_5614-5613 | PTS system | TTGGCACGCTAATTGCA | −387 | LevR |
| HD73_0179-0178 | Arginine and proline metabolism | TTGGTATGACAAAAGCA | −289 | Unknown |
| HD73_1649 | Lysine biosynthesis | TTGGAGATGTTGATGCG | −169 | Unknown |
| HD73_2025-2030 | Valine, leucine, and isoleucine metabolism | TCGGAGCATCGCTTGCG | −590 | Unknown |
| HD73_4943 | Acetate-CoA ligase | ATGGCTTAGAAAGAGCG | −166 | Unknown |
Distance between the −12 region of the promoter relative to the initiation codon.
Table 3.
Genes controlled by σ54 in B. thuringiensis HD73.
| Genes ID | Annotation | Fold-change |
|---|---|---|
| HD73_0366 | 4-aminobutyrate aminotransferase | 9.165 |
| HD73_0367 | Sensory box sigma-54 dependent DNA-binding response regulator | 2.860 |
| HD73_0368 | Succinic semialdehyde dehydrogenase | 2.410 |
| HD73_0369 | Quaternary ammonium compound-resistance protein | 2.377 |
| HD73_0560 | Biotin carboxyl carrier protein | - |
| HD73_0561 | Hypothetical protein | - |
| HD73_0562 | Amino-acid permease rocC | - |
| HD73_1024 | Proline racemase | 62.621 |
| HD73_1025 | Hypothetical protein | 3.687 |
| HD73_1070 | Glutamine amidotransferase, class I | 5.429 |
| HD73_2540 | L-lysine 2,3-aminomutase | 2.685 |
| HD73_2541 | Cytoplasmic protein | 2.296 |
| HD73_3140 | Hypothetical protein | 73.623 |
| HD73_3139 | Hypothetical protein | 80.805 |
| HD73_3138 | Sarcosine oxidase alpha subunit | 18.422 |
| HD73_3142 | Sarcosine oxidase, beta subunit | 35.033 |
| HD73_3143 | Proline racemase | 35.278 |
| HD73_3144 | Hypothetical protein | 31.999 |
| HD73_3145 | Dihydrodipicolinate synthase | 34.853 |
| HD73_3146 | Aldehyde dehydrogenase | 28.844 |
| HD73_3147 | Amino acid carrier protein | 37.362 |
| HD73_3213 | Acetoin:2,6-dichlorophenolindophenol oxidoreductase subunit alpha | 25.424 |
| HD73_3214 | TPP-dependent acetoin dehydrogenase E1 alpha-subunit | 19.979 |
| HD73_3215 | Acetoin:2,6-dichlorophenolindophenol oxidoreductase subunit beta | 9.610 |
| HD73_3216 | Dihydrolipoyllysine-residue acetyltransferase component of acetoin cleaving system | 9.326 |
| HD73_3217 | Dihydrolipoyl dehydrogenase | 4.337 |
| HD73_4161 | Proline dipeptidase | 103.525 |
| HD73_4468 | Phosphate butyryltransferase | 6.110 |
| HD73_4467 | Leucine dehydrogenase | - |
| HD73_4466 | Branched-chain-fatty-acid kinase | 2.728 |
| HD73_4465 | Dihydrolipoamide dehydrogenase | - |
| HD73_4464 | BfmBAa | - |
| HD73_4463 | 2-oxoisovalerate dehydrogenase subunit beta | - |
| HD73_4462 | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex | - |
| HD73_5327 | NADPH dehydrogenase, quinone | - |
| HD73_5613 | PTS system cellobiose-specific IIC component | 0.460 |
| HD73_5614 | PTS system cellobiose-specific IIC component | 0.460 |
| HD73_0179 | Pyrroline-5-carboxylate reductase | 2.125 |
| HD73_0178 | YtbE (Aldo/keto reductase YtbE) | 2.024 |
| HD73_1649 | Diaminopimelate decarboxylase | 2.270 |
| HD73_2025 | Branched-chain amino acid aminotransferase | 3.868 |
| HD73_2026 | Hypothetical protein | 4.825 |
| HD73_2027 | acetolactate synthase 1 regulatory subunit | 4.073 |
| HD73_2028 | Ketol-acid reductoisomerase | 4.921 |
| HD73_2029 | Dihydroxy-acid dehydratase | 3.666 |
| HD73_2030 | IlvA | 3.120 |
| HD73_4943 | Acetate-CoA ligase | 2.250 |
-, No detection in DNA microarray.
Prokaryotic EBPS encoded by the Bt genome
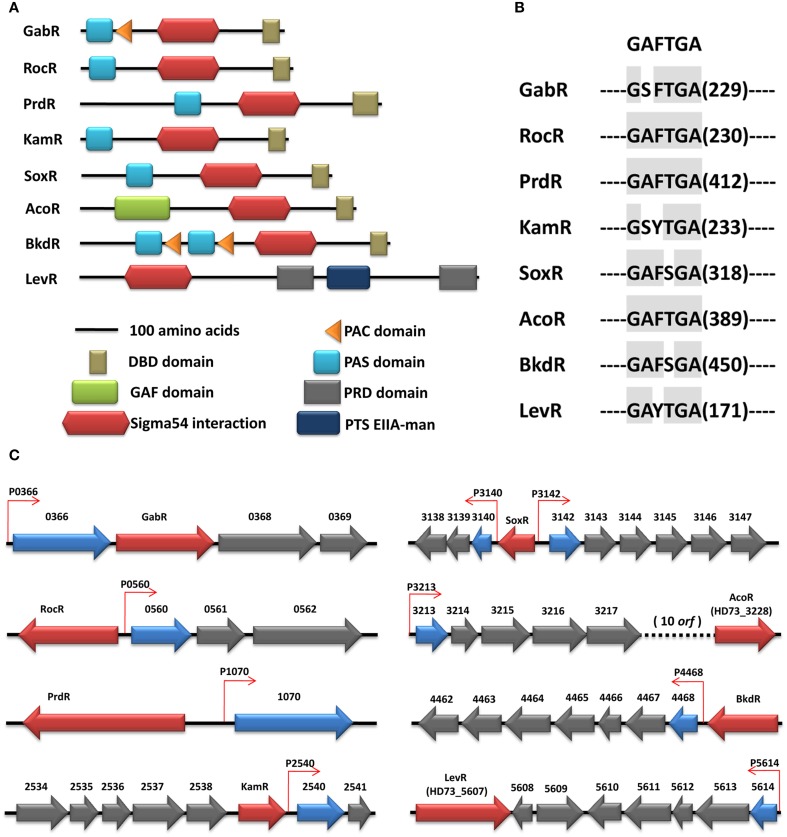
The activation of bEBPs is required for σ54-dependent transcription, and the identification of these proteins is therefore important in order to obtain a complete description of the σ54 regulon. Using hidden Markov models, eight bEBPs were identified in the HD73 genome with σ54 activator domains (Figure 3A). Four different N-terminal regulatory domain arrangements were identified in these eight proteins: those with a Per, ARNT, and Sim (PAS) domain (Ponting and Aravind, 1997), either alone or in combination with an aspartokinase, chorismate mutase, and TyrA (ACT or PAC) domain (Bush and Dixon, 2012); a cyclic GMP [cGMP]-specific and -stimulated phosphodiesterases, Anabaena adenylate cyclases, and E. coli FhlA (GAF) domain (Bush and Dixon, 2012); PTS regulatory domains (PRDs); and a PTS system fructose IIA component (PTS EIIA-man). PAS domains were identified in six σ54-dependent activators: HD73_0367 (GabR), HD73_0559 (RocR), HD73_1069 (PrdR), HD73_2539 (KamR), HD73_3141 (SoxR), and HD73_4469 (BkdR) (Figure 3A). GabR and BkdR also contained PAC domains; interestingly, the latter had two PAS/PAC domains, possibly for sensing different signals. Only one σ54-dependent activator, HD73_3228 (AcoR), harbored a GAF domain. Two alternative regulatory domains—a PRD domain and PTS EIIA-man—were present in HD73_5607 (LevR). Sequence alignments of bEBP AAA+ domains revealed a highly conserved GAFTGA motif (Zhang et al., 2009), in all eight putative bEBPs of Bt HD73 (Figure 3B), reflecting its importance for σ54- dependent transcription.
Figure 3.
(A) Domain structure of the EBPs in the Bt HD73 genome. The type of domains is indicated. All proteins are drawn at the scale indicated. (B) Conserved domain of the EBPs. The number indicates the distance between the conserved domain relative to the first amino acid. (C) Chromosomal context of EBPs in the Bt HD73 genome. The chromosomal context of those EBPs which have an associated σ54 promoter is shown here. Genes encoding EBPs are colored in red, whereas the genes or the first gene of the operons controlled by σ54 are represented in blue. The −12/−24 promoters are represented as small arrows and colored in red.
An important feature of σ54-dependent activators is their tendency to map close to their target promoters (Studholme, 2002). That was indeed the case for the majority of the Bt HD73 regulators, which had three typical organizations (Figure 3C). The most frequently observed was one in which the target promoter was located in the same direction as the activator (GabR, KamR, AcoR, and BkdR); alternatively, σ54-dependent promoters could be located immediately up- or downstream of the activator in the opposite orientation (e.g., RocR, PrdR, and LevR); in a third arrangement, the activator was located between two target promoters (e.g., SoxR).
Identification of EBP target genes
Several bEBP target genes were identified in the present work. We previously characterized the expression of the GABA pathway gene cluster in which gabT (HD73_0366) is controlled by σ54 and activated by GabR (Zhu et al., 2010), which specifically bound to a repeat region 58 bp upstream of the start codon (Peng et al., 2014). HD73_2540 (kamA, encoding l-lysine 2, 3-aminomutase) and its six upstream genes (HD73_2539–2534) form two operons. The activation of the kamA promoter—which is a typical −12/−24 sequence—was abolished in the ΔsigL mutant from T0 to T4 and was reduced from T4 to T7 as compared to the HD73 strain. The promoter was activated by KamR prior to T7 and controlled by σK and GerE after T10, the late stage of sporulation (Zhang et al., 2014).
The other eight typical −12/−24 promoters (P0560, P1024, P1070, P3140, P3142, P3213, P4468, and P5614) were analyzed to determine whether they are activated by their neighboring σ54-dependent activator. Mutants of six bEBPs (rocR, prdR, soxR, acoR, bkdR, and levR) were generated by homologous recombination and the expression of putative −12/−24 promoters was assessed with lacZ fusion constructs in each mutant background. The β-galactosidase assays showed that the activation of P0560, P1070, P3140, P3142, P3213, P4468, and P5614 was markedly decreased in the corresponding EBP mutants (rocR, prdR, soxR, acoR, bkdR, and levR, respectively), and these seven promoters were also controlled by σ54 (Figures 2A–D,F–H). Thus, RocR, PrdR, SoxR, AcoR, BkdR, and LevR are the EBPs of HD73_0560 (biotin carboxyl carrier protein), HD73_1070 (glutamine amidotransferase), HD73_3140 (hypothetical protein), HD73_3142 (sarcosine oxidase, β subunit), HD73_3213 (acetoin, 2, 6-dichlorophenolindophenol oxidoreductase subunit alpha), HD73_4468 (phosphate butyryltransferase), and HD73_5614 (PTS system cellobiose-specific IIC component), respectively. As an exception, the activation of P1024—which is also a typical −12/−24 promoter—was abolished in the ΔsigL but was unaltered in the prdR mutant, suggesting that PrdR is not the EBP of HD73_1024 (proline racemase).
Discussion
In this work, computational predictions combined with a functional analysis were used to investigate the Bt HD73 σ54 regulon. This regulon comprises five genes and 11 operons for a total of 47 genes, most of which possess typical −12/−24 promoters and are present in other B. cereus group strains, with the exception of HD73_1070 (glutamine amidotransferase) which is present in only a few strains such as FRI-35 (Additional file 3). These findings suggest that the σ54 regulon is conserved in the B. cereus group and provide insights into evolutionary relationships among its members, as well as mechanisms of metabolic regulation that contribute to host range. Most genes in the σ54 regulon are related to nitrogen- and carbon-metabolism pathways. Significantly, four operons (aco, bkd, lev, and roc) are involved in the acetoin catabolic pathway, isoleucine and valine utilization, generating sugar synthesis metabolites, and arginine catabolism, which is similar to B. subtilis in terms of putative function. These operons are controlled by σ54 and are regulated by the AcoR, BkdR, LevR, and RocR proteins both in Bt and B. subtilis (Debarbouille et al., 1991a, 1999; Ali et al., 2001, 2003). Interestingly, five other σ54-dependent genes or gene clusters, which are regulated by four EBPs (GabR, PrdR, KamR, and SoxR), have a unique organization in the B. cereus group. These σ54-dependent genes are involved in the GABA pathway (Zhu et al., 2010); glutamine, l-lysine (Zhang et al., 2014), and sarcosine metabolism; and are controlled by other σ70–type Sigma factors in some bacteria (Nishiya et al., 1998; Schneider et al., 2002; Errington, 2003; Belitsky and Sonenshein, 2004; Steil et al., 2005). The remaining seven σ54-dependent operons, which were not recognized by the corresponding EBPs, are involved in arginine and proline metabolism, lysine biosynthesis, branched-chain amino acids biosynthesis, ubiquinone- and other terpenoid-quinone biosynthesis reactions, and catabolism by acetate-CoA ligase. Our data reveals for the first time the seven operons mentioned above are controlled by σ54 in the B. cereus group.
σ54-mediated control of transcription is not only associated with nitrogen and carbon metabolism, but with a wider range of cellular processes and physiology in the bacteria. It was shown that its role also encompasses the regulation of flagellar biosynthesis in E. coli and Geobacter sulfurreducens (Leang et al., 2009; Zhao et al., 2010); osmotolerance in Listeria (Okada et al., 2008); and motility, biofilm formation, luminescence, and colonization in Vibrio fischeri (Wolfe et al., 2004; Visick, 2009). We were reported that transcription of the lysine-2, 3-aminomutase gene in the kam locus of Bt is controlled both by σ54 and the RNA polymerase sigma factor σK, which is associated with late-stage sporulation (Zhang et al., 2014). The mutation of kamR slightly decreased the sporulation rate, suggesting that the metabolic pathways regulated by KamR may be involve in sporulation (Zhang et al., 2014). Herein, data from the DNA microarray analysis revealed 255 genes that were downregulated and 121 that were upregulated in the ΔsigL mutant relative to the wild-type strain. Among the downregulated genes, 17 genes were associated with sporulation (Additional file 1) but lacked typical −12/−24 promoters, suggesting that σ54 may play a role in sporulation by controlling metabolic pathways in the B. cereus group and may indirectly regulate some sporulation related genes. These findings should open very interesting avenues of investigation in future studies.
The transcription of many σ54-controlled genes involved in physiological processes is induced or repressed by various environmental signals, which are mediated by the N-terminal domains of bEBPs and thereby regulate the activity of the central AAA+ domain (Bush and Dixon, 2012). For example, the regulatory domain of FhlA binds formate to activate transcription of the formate hydrogen lyase system in E. coli (Hopper and Bock, 1995). The acetoin catabolic pathway, which is activated by AcoR, is induced by acetoin in B. subtilis (Ali et al., 2001). The σ54-dependent bkd and roc operons were most strongly and specifically induced after ammonium starvation (Tam Le et al., 2007). Our previous studies also showed that the specific signaling factors GABA and succinic semialdehyde activated expression of gabT, which encodes the GABA transaminase of the GABA pathway. Its inducible activation is controlled by σ54 and regulated by GabR, and the PAS domain in GabR represses its enhancer transcriptional activity of the gabT promoter (Peng et al., 2014). However, in this study, the DNA microarray data was generated from cells grown in SSM medium without specific inducers of σ54-regulated promoters, containing only a nutrient broth with a low concentration of metal salts. Relative to the wild-type strain, the differentially expressed genes in the ΔsigL mutant reflect the basal gene expression levels. It will be interesting to compare the effects of sigL mutations and EBP mutations under inducing conditions in future work, which may reveal that additional σ54-dependent genes participate in physiological functions.
Author contributions
QP and FS designed the research. QP performed the experimental work and drafted the manuscript. QP constructed the promoter with lacZ fusions strain and carried out β-galactosidase assay. GW constructed the EBP mutants. GL participated in DNA microarray analysis. FS and JZ critically revised the manuscript for intellectual content. All authors read and approved the final version of the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (nos. 31270111 and 31300085). The authors thank Dr. Christina Nielsen-LeRoux and Dr. Didier Lereclus from the Institut National de la Recherche Agronomique, and Dr. Mario Soberon and Dr. Alejandra Bravo from Instituto de Biotecnologia, Universidad Nacional Autonoma de Mexico for their critical suggestions.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00407/abstract
References
- Agaisse H., Lereclus D. (1994). Structural and functional analysis of the promoter region involved in full expression of the cryIIIA toxin gene of Bacillus thuringiensis. Mol. Microbiol. 13, 97–107. 10.1111/j.1365-2958.1994.tb00405.x [DOI] [PubMed] [Google Scholar]
- Ali N. O., Bignon J., Rapoport G., Debarbouille M. (2001). Regulation of the acetoin catabolic pathway is controlled by sigma L in Bacillus subtilis. J. Bacteriol. 183, 2497–2504. 10.1128/JB.183.8.2497-2504.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali N. O., Jeusset J., Larquet E., Le Cam E., Belitsky B., Sonenshein A. L., et al. (2003). Specificity of the interaction of RocR with the rocG-rocA intergenic region in Bacillus subtilis. Microbiology 149, 739–750. 10.1099/mic.0.26013-0 [DOI] [PubMed] [Google Scholar]
- Arnaud M., Chastanet A., Debarbouille M. (2004). New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70, 6887–6891. 10.1128/AEM.70.11.6887-6891.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arous S., Buchrieser C., Folio P., Glaser P., Namane A., Hebraud M., et al. (2004). Global analysis of gene expression in an rpoN mutant of Listeria monocytogenes. Microbiology 150, 1581–1590. 10.1099/mic.0.26860-0 [DOI] [PubMed] [Google Scholar]
- Baev M. V., Baev D., Radek A. J., Campbell J. W. (2006). Growth of Escherichia coli MG1655 on LB medium: monitoring utilization of amino acids, peptides, and nucleotides with transcriptional microarrays. Appl. Microbiol. Biotechnol. 71, 317–322. 10.1007/s00253-005-0310-5 [DOI] [PubMed] [Google Scholar]
- Belitsky B. R., Sonenshein A. L. (2004). Modulation of activity of Bacillus subtilis regulatory proteins GltC and TnrA by glutamate dehydrogenase. J. Bacteriol. 186, 3399–3407. 10.1128/JB.186.11.3399-3407.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M., Gallegos M. T., Studholme D. J., Guo Y., Gralla J. D. (2000). The bacterial enhancer-dependent sigma(54) (sigma(N)) transcription factor. J. Bacteriol. 182, 4129–4136. 10.1128/JB.182.15.4129-4136.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush M., Dixon R. (2012). The role of bacterial enhancer binding proteins as specialized activators of sigma54-dependent transcription. Microbiol. Mol. Biol. Rev. 76, 497–529. 10.1128/MMBR.00006-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldara M., Minh P. N., Bostoen S., Massant J., Charlier D. (2007). ArgR-dependent repression of arginine and histidine transport genes in Escherichia coli K-12. J. Mol. Biol. 373, 251–267. 10.1016/j.jmb.2007.08.013 [DOI] [PubMed] [Google Scholar]
- Cases I., Ussery D. W., de Lorenzo V. (2003). The sigma54 regulon (sigmulon) of Pseudomonas putida. Environ. Microbiol. 5, 1281–1293. 10.1111/j.1462-2920.2003.00528.x [DOI] [PubMed] [Google Scholar]
- da Silva Neto J. F., Koide T., Gomes S. L., Marques M. V. (2010). Global gene expression under nitrogen starvation in Xylella fastidiosa: contribution of the sigma54 regulon. BMC Microbiol. 10:231. 10.1186/1471-2180-10-231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debarbouille M., Gardan R., Arnaud M., Rapoport G. (1999). Role of bkdR, a transcriptional activator of the sigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J. Bacteriol. 181, 2059–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debarbouille M., Martin-Verstraete I., Klier A., Rapoport G. (1991a). The transcriptional regulator LevR of Bacillus subtilis has domains homologous to both sigma 54- and phosphotransferase system-dependent regulators. Proc. Natl. Acad. Sci. U.S.A. 88, 2212–2216. 10.1073/pnas.88.6.2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debarbouille M., Martin-Verstraete I., Kunst F., Rapoport G. (1991b). The Bacillus subtilis sigL gene encodes an equivalent of sigma 54 from gram-negative bacteria. Proc. Natl. Acad. Sci. U.S.A. 88, 9092–9096. 10.1073/pnas.88.20.9092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B., Marchal K., Vanderleyden J., Michiels J. (2002). Prediction and overview of the RpoN-regulon in closely related species of the Rhizobiales. Genome Biol. 3:RESEARCH0076. 10.1186/gb-2002-3-12-research0076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong T. G., Mekalanos J. J. (2012). Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res. 40, 7766–7775. 10.1093/nar/gks567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J. (2003). Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1, 117–126. 10.1038/nrmicro750 [DOI] [PubMed] [Google Scholar]
- Francke C., Groot Kormelink T., Hagemeijer Y., Overmars L., Sluijter V., Moezelaar R., et al. (2011). Comparative analyses imply that the enigmatic Sigma factor 54 is a central controller of the bacterial exterior. BMC Genomics 12:385. 10.1186/1471-2164-12-385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerout-Fleury A. M., Shazand K., Frandsen N., Stragier P. (1995). Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167, 335–336. 10.1016/0378-1119(95)00652-4 [DOI] [PubMed] [Google Scholar]
- Han C. S., Xie G., Challacombe J. F., Altherr M. R., Bhotika S. S., Brown N., et al. (2006). Pathogenomic sequence analysis of Bacillus cereus and Bacillus thuringiensis isolates closely related to Bacillus anthracis. J. Bacteriol. 188, 3382–3390. 10.1128/JB.188.9.3382-3390.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser F., Pessi G., Friberg M., Weber C., Rusca N., Lindemann A., et al. (2007). Dissection of the Bradyrhizobium japonicum NifA+sigma54 regulon, and identification of a ferredoxin gene (fdxN) for symbiotic nitrogen fixation. Mol. Genet. Genomics 278, 255–271. 10.1007/s00438-007-0246-9 [DOI] [PubMed] [Google Scholar]
- Heidrich N., Chinali A., Gerth U., Brantl S. (2006). The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism. Mol. Microbiol. 62, 520–536. 10.1111/j.1365-2958.2006.05384.x [DOI] [PubMed] [Google Scholar]
- Helgason E., Okstad O. A., Caugant D. A., Johansen H. A., Fouet A., Mock M., et al. (2000). Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66, 2627–2630. 10.1128/AEM.66.6.2627-2630.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper S., Bock A. (1995). Effector-mediated stimulation of ATPase activity by the sigma 54-dependent transcriptional activator FHLA from Escherichia coli. J. Bacteriol. 177, 2798–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiupakis A. K., Reitzer L. (2002). ArgR-independent induction and ArgR-dependent superinduction of the astCADBE operon in Escherichia coli. J. Bacteriol. 184, 2940–2950. 10.1128/JB.184.11.2940-2950.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leang C., Krushkal J., Ueki T., Puljic M., Sun J., Juarez K., et al. (2009). Genome-wide analysis of the RpoN regulon in Geobacter sulfurreducens. BMC Genomics 10:331. 10.1186/1471-2164-10-331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. K., Newman J. D., Keasling J. D. (2005). Catabolite repression of the propionate catabolic genes in Escherichia coli and Salmonella enterica: evidence for involvement of the cyclic AMP receptor protein. J. Bacteriol. 187, 2793–2800. 10.1128/JB.187.8.2793-2800.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lereclus D., Arantes O., Chaufaux J., Lecadet M. (1989). Transformation and expression of a cloned delta-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol. Lett. 51, 211–217. 10.1016/0378-1097(89)90511-9 [DOI] [PubMed] [Google Scholar]
- Magasanik B. (1989). Regulation of transcription of the glnALG operon of Escherichia coli by protein phosphorylation. Biochimie 71, 1005–1012. 10.1016/0300-9084(89)90104-1 [DOI] [PubMed] [Google Scholar]
- Matta M. K., Lioliou E. E., Panagiotidis C. H., Kyriakidis D. A., Panagiotidis C. A. (2007). Interactions of the antizyme AtoC with regulatory elements of the Escherichia coli atoDAEB operon. J. Bacteriol. 189, 6324–6332. 10.1128/JB.00214-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick M. J. (1993). In a class of its own—the RNA polymerase sigma factor sigma 54 (sigma N). Mol. Microbiol. 10, 903–909. 10.1111/j.1365-2958.1993.tb00961.x [DOI] [PubMed] [Google Scholar]
- Nishiya Y., Toda A., Imanaka T. (1998). Gene cluster for creatinine degradation in Arthrobacter sp. TE1826. Mol. Gen. Genet. 257, 581–586. 10.1007/s004380050685 [DOI] [PubMed] [Google Scholar]
- Okada Y., Makino S., Okada N., Asakura H., Yamamoto S., Igimi S. (2008). Identification and analysis of the osmotolerance associated genes in Listeria monocytogenes. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess 25, 1089–1094. 10.1080/02652030802056634 [DOI] [PubMed] [Google Scholar]
- Peng Q., Yang M., Wang W., Han L., Wang G., Wang P., et al. (2014). Activation of gab cluster transcription in Bacillus thuringiensis by gamma-aminobutyric acid or succinic semialdehyde is mediated by the Sigma 54-dependent transcriptional activator GabR. BMC Microbiol. 14:306. 10.1186/s12866-014-0306-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perchat S., Dubois T., Zouhir S., Gominet M., Poncet S., Lemy C., et al. (2011). A cell-cell communication system regulates protease production during sporulation in bacteria of the Bacillus cereus group. Mol. Microbiol. 82, 619–633. 10.1111/j.1365-2958.2011.07839.x [DOI] [PubMed] [Google Scholar]
- Ponting C. P., Aravind L. (1997). PAS: a multifunctional domain family comes to light. Curr. Biol. 7, R674–R677. 10.1016/S0960-9822(06)00352-6 [DOI] [PubMed] [Google Scholar]
- Reitzer L., Schneider B. L. (2001). Metabolic context and possible physiological themes of sigma(54)-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 65, 422–444. 10.1128/MMBR.65.3.422-444.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer P., Millet J., Aubert J. P. (1965). Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. U.S.A. 54, 704–711. 10.1073/pnas.54.3.704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider B. L., Ruback S., Kiupakis A. K., Kasbarian H., Pybus C., Reitzer L. (2002). The Escherichia coli gabDTPC operon: specific gamma-aminobutyrate catabolism and nonspecific induction. J. Bacteriol. 184, 6976–6986. 10.1128/JB.184.24.6976-6986.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepf E., Crickmore N., Van Rie J., Lereclus D., Baum J., Feitelson J., et al. (1998). Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62, 775–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steil L., Serrano M., Henriques A. O., Volker U. (2005). Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology 151, 399–420. 10.1099/mic.0.27493-0 [DOI] [PubMed] [Google Scholar]
- Studholme D. J. (2002). Enhancer-dependent transcription in Salmonella enterica Typhimurium: new members of the sigmaN regulon inferred from protein sequence homology and predicted promoter sites. J. Mol. Microbiol. Biotechnol. 4, 367–374. [PubMed] [Google Scholar]
- Tam Le T., Eymann C., Antelmann H., Albrecht D., Hecker M. (2007). Global gene expression profiling of Bacillus subtilis in response to ammonium and tryptophan starvation as revealed by transcriptome and proteome analysis. J. Mol. Microbiol. Biotechnol. 12, 121–130. 10.1159/000096467 [DOI] [PubMed] [Google Scholar]
- Visick K. L. (2009). An intricate network of regulators controls biofilm formation and colonization by Vibrio fischeri. Mol. Microbiol. 74, 782–789. 10.1111/j.1365-2958.2009.06899.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Zhang J., Song F., Wu J., Feng S., Huang D. (2006). Engineered Bacillus thuringiensis GO33A with broad insecticidal activity against lepidopteran and coleopteran pests. Appl. Microbiol. Biotechnol. 72, 924–930. 10.1007/s00253-006-0390-x [DOI] [PubMed] [Google Scholar]
- Wolfe A. J., Millikan D. S., Campbell J. M., Visick K. L. (2004). Vibrio fischeri sigma54 controls motility, biofilm formation, luminescence, and colonization. Appl. Environ. Microbiol. 70, 2520–2524. 10.1128/AEM.70.4.2520-2524.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Furusawa C., Nagahisa K., Kashiwagi A., Yomo T., Shimizu H. (2007). Analysis of fluctuation in protein abundance without promoter regulation based on Escherichia coli continuous culture. Biosystems 90, 614–622. 10.1016/j.biosystems.2007.02.001 [DOI] [PubMed] [Google Scholar]
- Zhang N., Joly N., Burrows P. C., Jovanovic M., Wigneshweraraj S. R., Buck M. (2009). The role of the conserved phenylalanine in the sigma54-interacting GAFTGA motif of bacterial enhancer binding proteins. Nucleic Acids Res. 37, 5981–5992. 10.1093/nar/gkp658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Yang M., Peng Q., Wang G., Zheng Q., Zhang J., et al. (2014). Transcription of the Lysine-2,3-Aminomutase Gene in the kam Locus of Bacillus thuringiensis subsp. kurstaki HD73 Is Controlled by Both sigma54 and sigmaK Factors. J. Bacteriol. 196, 2934–2943. 10.1128/JB.01675-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Liu M., Burgess R. R. (2010). Promoter and regulon analysis of nitrogen assimilation factor, sigma54, reveal alternative strategy for E. coli MG1655 flagellar biosynthesis. Nucleic Acids Res. 38, 1273–1283. 10.1093/nar/gkp1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Peng Q., Song F., Jiang Y., Sun C., Zhang J., et al. (2010). Structure and regulation of the gab gene cluster, involved in the gamma-aminobutyric acid shunt, are controlled by a sigma54 factor in Bacillus thuringiensis. J. Bacteriol. 192, 346–355. 10.1128/JB.01038-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.