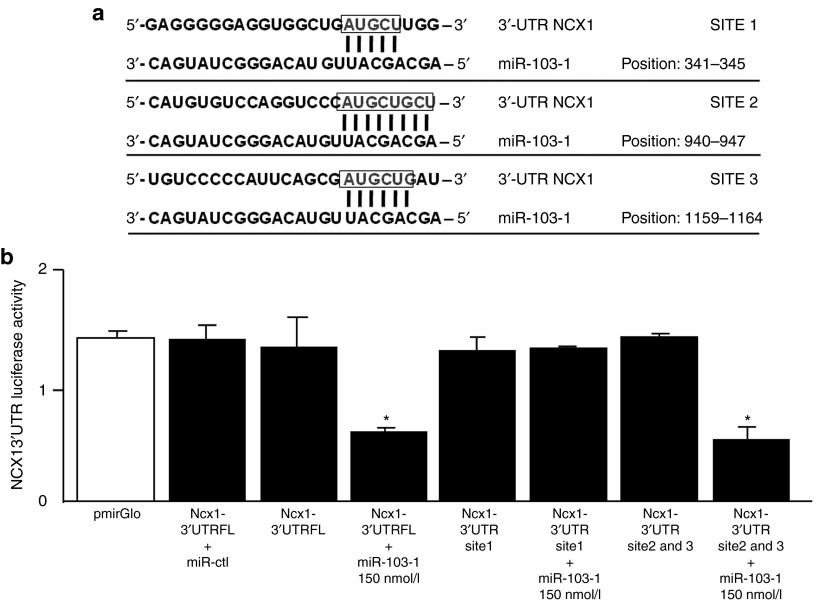
Figure 1.
miR-103-1 regulates NCX1 by targeting 3′-UTR of Ncx1 mRNA. (a) Predicted miR-103-1 binding sites in the 3′-UTRs of NCX1 mRNA. NCX1 contains 3 sites of annealing with candidate miRNA. Site position relative to the beginning of the 3′-UTRs is indicated. Target sequences are highlighted by a box, and base pairing between miR-103-1 and the target site is marked by vertical lines. The most relevant sites of miRNA-mRNA interaction (8 bp of interaction) relatively to the beginning of the 3′-UTRs region of NCX1 (rattus norvegicus) is shown. (b) Luciferase activity assay has been evaluated in PC12 cells by coexpressing the vector cointaining 3′ UTRs of NCX1 and candidate miRNA (miR-103-1). The activity of luciferase enzyme was normalized to β-galactosidase activity and compared with empty vector measurements (pmiR empty). This experiment was performed in triplicate and is representative of three independent experiments. Results show that a strong decrease in luciferase activity is observed when the pmirGLO vector, containing sites 2 and 3 out of three total interaction sites, was cotransfected with mirR-103-1 at 150 nmol/l in PC12 cells. The panel shows that site 1 is ineffective in NCX1 mRNA modulation by miRNA. Data are expressed as mean ± SEM. *P < 0.01.