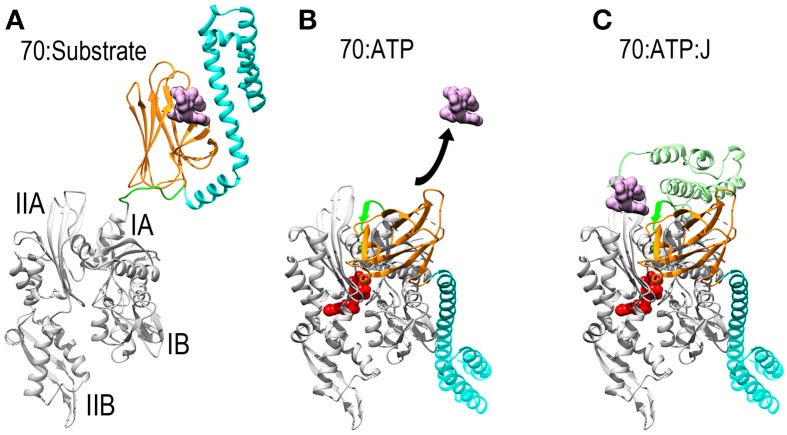
Figure 1.
Mechanisms of ATP dependent protein substrate release and binding by Hsp70. (A) Ribbon model of Hsp70 ADP state structure [pbd 2KHO (Bertelsen et al., 2009)] with the NBD in gray, PBD-β and PBD-α in orange and cyan, respectively, and the linker between the NBD and PBD in green. A space-filling model of substrate peptide (magenta) is shown bound to the PBD. (B) Model of Hsp70 ATP state structure [pdb 4B9Q (Kityk et al., 2012)]. Binding of ATP induces NBD to close around the nucleotide which expands the groove between subdomains IA and IIA, allowing the interdomain linker to bind in this groove. This creates a binding site for PBD-β, which separates from PBD-α and releases the protein substrate. (C): Hsp70:ATP:J domain structure [modeled from pdb 2QWQ (Jiang et al., 2007)]. The J domain of a J protein (pale green) binds to the IIA-Linker-IA surface induced by ATP binding and its distinctive substrate binding domain binds and present a protein substrate to Hsp70. Upon ATP hydrolysis the J protein releases Hsp70 which clamps down on the protein substrate to return to the conformation shown in (A).