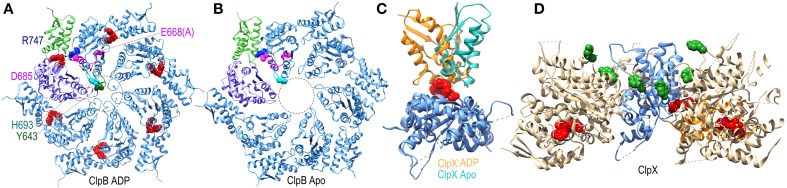
Figure 7.
Nucleotide dependent conformational changes underlying protein threading through the ClpB/Hsp104 pore. (A) A ClpB D2 hexamer ring assembled from the structure of ClpB D2 with ADP (red) bound [pdb 4FCV (Biter et al., 2012)]. The small and large subdomains of one of the D2 domains are colored green and purple, respectively, to illustrate how nucleotide binds at the interface of these two subdomains. ADP bound to this domain is sensed by the arginine finger (R747 in blue) of the adjacent domain. R747 contacts D685 of the ISS motif (magenta). Also in magenta are the proximal D9 helix and E668 (mutated to ala in this structure) of the Walker B motif which contacts bound ADP. These residues sit at the base of a β-hairpin which contains H693 at its tip, and which is packed against the pore loop with its conserved tyrosine (Y643 in green). The pore loops lie within the area defined by the dotted circle and are ordered in the ADP-bound structure. (B) Apo ClpB D2 hexamer ring [pdb 4FCT (Biter et al., 2012)]. The pore loops are disordered and not visible in the apo state. (C) Superimposed structures of apo and ADP-bound ClpX [pdb 3HWS (Glynn et al., 2009)] protomers show how ADP binding induces large rotations between the large and small subdomains. (D) Structure of an asymmetric ClpX hexamer illustrates how the ADP induced changes in the orientation between large and small subdomains shift the positions of the protomers and of the substrate-binding pore loops (highlighted by the green space-filling renderings of the conserved tyrosines at the tips of the loops) in the hexamer (the large and small subdomains of one of the protomers are colored, respectively, blue and orange).