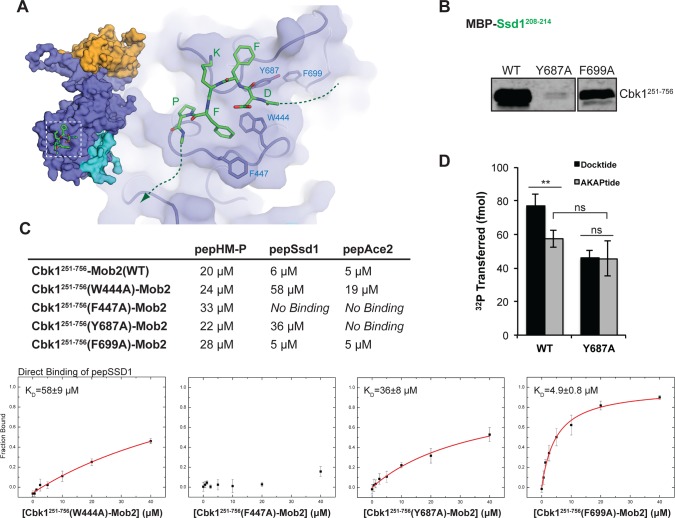
Fig 5. Structural model of the Cbk1–docking peptide complex.
(A) AutoDock binding simulation of Ssd1 docking fragment (DFKFP) identifies an interaction surface on Cbk1 and suggests important aromatic interactions with residues W444, F447, and Y687. (B) Y687A but not nearby F699A abrogate Cbk1 pulldown by Ssd1208–214 as predicted by the model. (C) FP analysis of putative docking motif binding site residues. The binding of all mutants to pepHM-P was similar to that of WT Cbk1, indicating that the mutants are folded properly (see first column in the table summarizing peptide binding affinities). W444A, F447A, and Y687A were reduced in their binding affinity for Ssd1 and Ace2 docking peptides, while F699A was not. (D) Peptide kinase assay of WT and Y687A mutant of Cbk1251–756–Mob2. Peptides are based on Ssd1 (210–229), which contains the N-terminal docking motif (Docktide: FKFP, AKAPtide: AKAP) and a Cbk1 consensus site. The presence of the WT docking motif enhances phosphorylation by WT Cbk1 relative to a peptide containing a mutant docking motif (similar to Fig 4F). Cbk1(Y687A) loses this enhancement when unable to bind WT docking motif. Data is plotted as mean ± standard error of the mean. Student’s unpaired t-test: **p < 0.01; ns, not significant. Data for (C) can be found S2 Data, and data for (D) can be found in S3 Data.