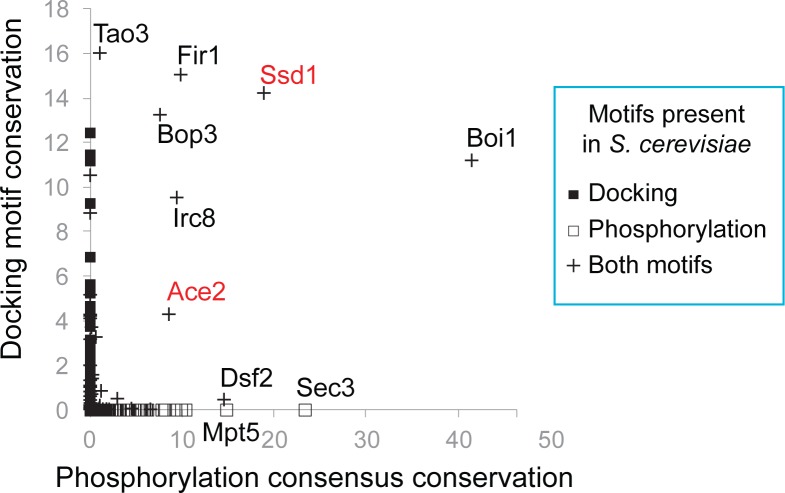
Fig 7. Co-conservation of docking and phosphorylation sites identifies known and likely Cbk1 regulatory targets.
Conservation of Cbk1 docking motifs and phosphorylation consensus sites in individual proteins, calculated using ConDens [48]. For each motif type, the most significant score among the matches in each protein is assigned as the score for that protein. The x-axis plots conservation scores of [YF]xFP sequences (Cbk1 docking), and the y-axis plots conservation scores of Hx[RK]xx[ST] sequences (Cbk1 phosphorylation consensus). The known Cbk1 substrates Ssd1 and Ace2 are highlighted in red. Proteins without docking motifs that score very highly for phosphorylation consensus conservation are noted. ConDens scores can found in S6 Data.