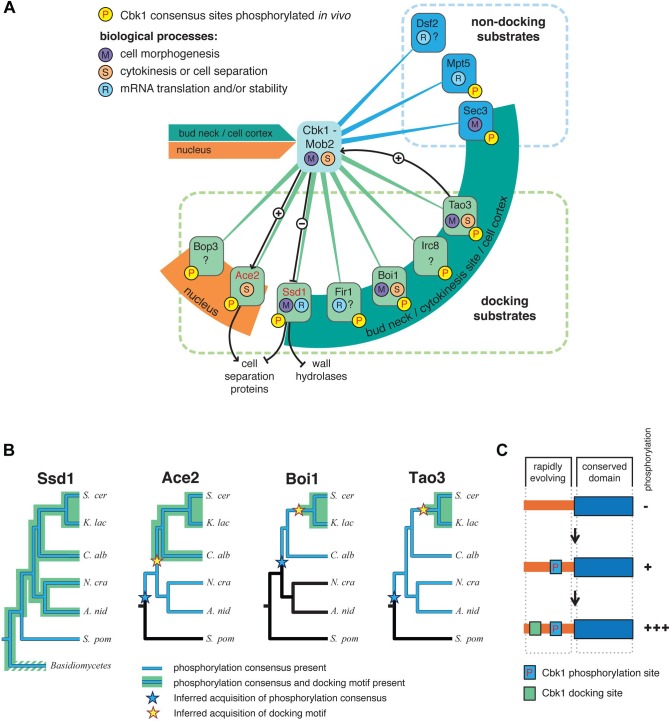
Fig 9. A probable network of proteins controlled by the yeast RAM network, and sequential evolution of docking in Cbk1 substrates.
(A) An outline of known and likely Cbk1 substrates in which the core docking motif is conserved (green lines) and likely substrates that lack a docking motif (blue lines). Ace2 and Ssd1 (red type) are known Cbk1 phosphorylation targets, and black lines indicate regulatory interactions inferred from molecular genetic analysis. Orange and green arcs indicate localization of target proteins to either the nucleus or sites of cell growth and cortical remodeling (bud neck/cytokinesis site/cell cortex): Cbk1 displays both of these localization patterns. (B) Distribution of phosphorylation consensus sites and docking motifs in known and likely Cbk1 substrates. Orthologs of Ace2, Boi1, and Tao3 are present in the lineages drawn. Blue indicates lineages in which the proteins have conserved phosphorylation consensus sites, while in those drawn in black, consensus sites are absent. Inferred acquisition of consensus sites is represented by a blue star. Lineages in which the proteins have conserved Cbk1 docking motifs are highlighted green, with inferred acquisition of this motif labeled by a yellow star. (C) Sequential addition model for acquisition of regulatory motifs during signaling system evolution. Substrates first become phosphorylation targets of a given upstream kinase. Docking motifs are subsequently acquired in unstructured, rapidly evolving sequence. The robustness of substrate phosphorylation increases upon docking motif acquisition. A. nid, Aspergillus nidulans; C. alb, Candida albicans; K. lac, Kluyveromyces lactis; N. cra, Neurospora crassa; S. cer, Saccharomyces cerevisiae; S. pom, S chizosaccharomyces pombe.